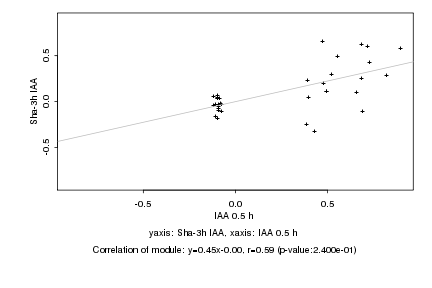
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IAA 0.5 h |
Log2 signal ratio
Sha-3h IAA |
| 1 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
0.894 |
0.582 |
| 2 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.819 |
0.285 |
| 3 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.723 |
0.434 |
| 4 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.716 |
0.600 |
| 5 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.688 |
-0.103 |
| 6 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.680 |
0.251 |
| 7 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.679 |
0.632 |
| 8 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
0.654 |
0.109 |
| 9 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.553 |
0.498 |
| 10 |
248801_at |
AT5G47370
|
HAT2 |
0.520 |
0.298 |
| 11 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.491 |
0.111 |
| 12 |
263935_at |
AT2G35930
|
AtPUB23, PUB23, plant U-box 23 |
0.490 |
0.118 |
| 13 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.477 |
0.204 |
| 14 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.470 |
0.655 |
| 15 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
0.425 |
-0.321 |
| 16 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.393 |
0.048 |
| 17 |
263931_at |
AT2G36220
|
unknown |
0.387 |
0.238 |
| 18 |
245041_at |
AT2G26530
|
AR781 |
0.385 |
-0.245 |
| 19 |
245563_at |
AT4G14580
|
CIPK4, CBL-interacting protein kinase 4, SnRK3.3, SNF1-RELATED PROTEIN KINASE 3.3 |
-0.124 |
0.058 |
| 20 |
245341_at |
AT4G16447
|
unknown |
-0.123 |
-0.042 |
| 21 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.112 |
-0.160 |
| 22 |
249388_at |
AT5G40090
|
CHL1, CHS1-like 1 |
-0.109 |
-0.025 |
| 23 |
259963_at |
AT1G53660
|
[nodulin MtN21 /EamA-like transporter family protein] |
-0.102 |
0.034 |
| 24 |
248847_at |
AT5G46510
|
VICTL, VARIATION IN COMPOUND TRIGGERED ROOT growth response-like |
-0.101 |
0.039 |
| 25 |
263276_at |
AT2G14100
|
CYP705A13, cytochrome P450, family 705, subfamily A, polypeptide 13 |
-0.099 |
0.074 |
| 26 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
-0.098 |
-0.185 |
| 27 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
-0.098 |
0.048 |
| 28 |
260631_at |
AT1G62350
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.097 |
-0.053 |
| 29 |
262972_at |
AT1G75620
|
[glyoxal oxidase-related protein] |
-0.096 |
-0.023 |
| 30 |
259365_at |
AT1G13300
|
HRS1, HYPERSENSITIVITY TO LOW PI-ELICITED PRIMARY ROOT SHORTENING 1 |
-0.095 |
-0.090 |
| 31 |
258172_at |
AT3G21620
|
[ERD (early-responsive to dehydration stress) family protein] |
-0.093 |
-0.069 |
| 32 |
247335_at |
AT5G63630
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.087 |
0.041 |
| 33 |
251214_at |
AT3G62500
|
unknown |
-0.083 |
-0.014 |
| 34 |
267416_at |
AT2G34980
|
SETH1 |
-0.080 |
-0.099 |
| 35 |
265537_at |
AT2G15870
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-0.078 |
-0.031 |