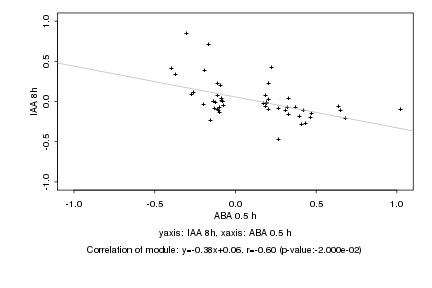
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA 0.5 h |
Log2 signal ratio
IAA 8h |
| 1 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
1.020 |
-0.087 |
| 2 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.678 |
-0.199 |
| 3 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.647 |
-0.107 |
| 4 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
0.633 |
-0.051 |
| 5 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.471 |
-0.143 |
| 6 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
0.463 |
-0.198 |
| 7 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.431 |
-0.261 |
| 8 |
259231_at |
AT3G11410
|
AHG3, ATPP2CA, ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA, PP2CA, protein phosphatase 2CA |
0.419 |
-0.100 |
| 9 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.403 |
-0.282 |
| 10 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.395 |
-0.183 |
| 11 |
265276_at |
AT2G28400
|
unknown |
0.371 |
-0.072 |
| 12 |
252660_at |
AT3G44440
|
unknown |
0.327 |
0.041 |
| 13 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.324 |
-0.157 |
| 14 |
253155_at |
AT4G35720
|
unknown |
0.319 |
-0.062 |
| 15 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.310 |
-0.111 |
| 16 |
248209_at |
AT5G53990
|
[UDP-Glycosyltransferase superfamily protein] |
0.263 |
-0.460 |
| 17 |
259793_at |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
0.262 |
-0.076 |
| 18 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.219 |
0.427 |
| 19 |
265553_at |
AT2G07550
|
[copia-like retrotransposon family, has a 4.7e-313 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
0.204 |
0.035 |
| 20 |
259012_at |
AT3G07360
|
ATPUB9, ARABIDOPSIS THALIANA PLANT U-BOX 9, PUB9, plant U-box 9 |
0.202 |
-0.099 |
| 21 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.202 |
0.226 |
| 22 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.192 |
-0.018 |
| 23 |
252193_at |
AT3G50060
|
MYB77, myb domain protein 77 |
0.189 |
-0.006 |
| 24 |
258936_s_at |
AT3G10100
|
ASY2 |
0.182 |
-0.060 |
| 25 |
247236_at |
AT5G64590
|
unknown |
0.182 |
0.086 |
| 26 |
250195_at |
AT5G14560
|
unknown |
0.170 |
-0.014 |
| 27 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.397 |
0.411 |
| 28 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.377 |
0.342 |
| 29 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
-0.308 |
0.856 |
| 30 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
-0.273 |
0.093 |
| 31 |
259502_at |
AT1G15670
|
KFB01, Kelch repeat F-box 1, KMD2, KISS ME DEADLY 2 |
-0.260 |
0.119 |
| 32 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.201 |
-0.028 |
| 33 |
265028_at |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.194 |
0.387 |
| 34 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.171 |
0.711 |
| 35 |
254408_at |
AT4G21390
|
B120 |
-0.157 |
-0.231 |
| 36 |
256784_at |
AT3G13674
|
unknown |
-0.142 |
0.003 |
| 37 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.136 |
0.005 |
| 38 |
258935_at |
AT3G10120
|
unknown |
-0.132 |
-0.083 |
| 39 |
249493_at |
AT5G39080
|
[HXXXD-type acyl-transferase family protein] |
-0.129 |
-0.010 |
| 40 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
-0.115 |
-0.095 |
| 41 |
254409_at |
AT4G21400
|
CRK28, cysteine-rich RLK (RECEPTOR-like protein kinase) 28 |
-0.113 |
0.226 |
| 42 |
261700_at |
AT1G32690
|
unknown |
-0.113 |
0.078 |
| 43 |
246826_at |
AT5G26310
|
UGT72E3 |
-0.107 |
-0.102 |
| 44 |
266721_at |
AT2G03220
|
ATFT1, ARABIDOPSIS THALIANA FUCOSYLTRANSFERASE 1, ATFUT1, ARABIDOPSIS THALIANA FUCOSYLTRANSFERASE 1, FT1, fucosyltransferase 1, MUR2, MURUS 2 |
-0.100 |
-0.072 |
| 45 |
257076_at |
AT3G19680
|
unknown |
-0.099 |
-0.136 |
| 46 |
262114_at |
AT1G02860
|
BAH1, BENZOIC ACID HYPERSENSITIVE 1, NLA, nitrogen limitation adaptation |
-0.093 |
0.208 |
| 47 |
260882_at |
AT1G29280
|
ATWRKY65, WRKY DNA-BINDING PROTEIN 65, WRKY65, WRKY DNA-binding protein 65 |
-0.089 |
0.025 |
| 48 |
252593_at |
AT3G45590
|
ATSEN1, splicing endonuclease 1, SEN1, splicing endonuclease 1 |
-0.089 |
0.046 |
| 49 |
256637_at |
AT3G12030
|
unknown |
-0.086 |
0.009 |
| 50 |
267624_at |
AT2G39660
|
BIK1, botrytis-induced kinase1 |
-0.086 |
0.003 |
| 51 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.078 |
-0.046 |