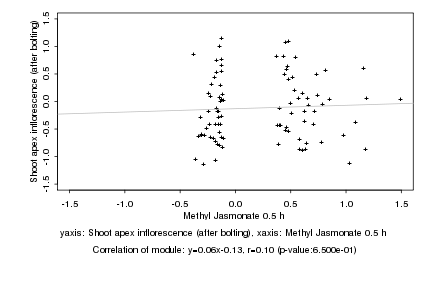
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Methyl Jasmonate 0.5 h |
Log2 signal ratio
Shoot apex inflorescence (after bolting) |
| 1 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
1.489 |
0.044 |
| 2 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
1.181 |
0.063 |
| 3 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
1.175 |
-0.856 |
| 4 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
1.157 |
0.615 |
| 5 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
1.083 |
-0.377 |
| 6 |
245209_at |
AT5G12340
|
unknown |
1.031 |
-1.125 |
| 7 |
253259_at |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
0.977 |
-0.599 |
| 8 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.843 |
0.048 |
| 9 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.807 |
0.572 |
| 10 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.787 |
-0.045 |
| 11 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.773 |
-0.726 |
| 12 |
245252_at |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
0.740 |
0.121 |
| 13 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
0.732 |
0.498 |
| 14 |
248793_at |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
0.706 |
-0.171 |
| 15 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
0.704 |
-0.412 |
| 16 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.652 |
-0.063 |
| 17 |
250149_at |
AT5G14700
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.644 |
0.068 |
| 18 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.642 |
-0.747 |
| 19 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.628 |
-0.863 |
| 20 |
245244_at |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
0.620 |
-0.359 |
| 21 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.616 |
-0.164 |
| 22 |
262171_at |
AT1G74950
|
JAZ2, JASMONATE-ZIM-DOMAIN PROTEIN 2, TIFY10B |
0.605 |
0.163 |
| 23 |
248794_at |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
0.602 |
-0.881 |
| 24 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
0.579 |
-0.672 |
| 25 |
247026_at |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
0.575 |
-0.869 |
| 26 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
0.570 |
0.070 |
| 27 |
259518_at |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
0.540 |
0.805 |
| 28 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.526 |
0.206 |
| 29 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.510 |
0.437 |
| 30 |
245041_at |
AT2G26530
|
AR781 |
0.503 |
-0.202 |
| 31 |
264083_at |
AT2G31230
|
ATERF15, ethylene-responsive element binding factor 15, ERF15, ethylene-responsive element binding factor 15 |
0.489 |
-0.027 |
| 32 |
259725_at |
AT1G61065
|
unknown |
0.478 |
-0.542 |
| 33 |
260429_at |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
0.475 |
0.400 |
| 34 |
265530_at |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
0.473 |
1.103 |
| 35 |
248293_at |
AT5G53050
|
[alpha/beta-Hydrolases superfamily protein] |
0.466 |
0.638 |
| 36 |
263972_at |
AT2G42760
|
unknown |
0.458 |
0.592 |
| 37 |
258189_at |
AT3G17860
|
JAI3, JASMONATE-INSENSITIVE 3, JAZ3, jasmonate-zim-domain protein 3, TIFY6B |
0.458 |
-0.461 |
| 38 |
255132_at |
AT4G08170
|
[Inositol 1,3,4-trisphosphate 5/6-kinase family protein] |
0.450 |
1.078 |
| 39 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.445 |
-0.515 |
| 40 |
263150_at |
AT1G54050
|
[HSP20-like chaperones superfamily protein] |
0.438 |
0.502 |
| 41 |
265680_at |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.434 |
0.825 |
| 42 |
259568_at |
AT1G20490
|
[AMP-dependent synthetase and ligase family protein] |
0.404 |
-0.425 |
| 43 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
0.396 |
-0.434 |
| 44 |
252679_at |
AT3G44260
|
AtCAF1a, CCR4- associated factor 1a, CAF1a, CCR4- associated factor 1a |
0.390 |
-0.109 |
| 45 |
248509_at |
AT5G50335
|
unknown |
0.388 |
-0.779 |
| 46 |
254651_at |
AT4G18160
|
ATKCO6, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 6, ATTPK3, KCO6, Ca2+ activated outward rectifying K+ channel 6, TPK3 |
0.373 |
-0.432 |
| 47 |
256457_at |
AT1G75230
|
[DNA glycosylase superfamily protein] |
0.363 |
0.819 |
| 48 |
245668_at |
AT1G28330
|
AtDRM1, DRM1, DORMANCY-ASSOCIATED PROTEIN 1, DYL1, dormancy-associated protein-like 1 |
-0.386 |
0.867 |
| 49 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.370 |
-1.038 |
| 50 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.335 |
-0.632 |
| 51 |
253165_at |
AT4G35320
|
unknown |
-0.322 |
-0.281 |
| 52 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
-0.314 |
-0.591 |
| 53 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.311 |
-0.600 |
| 54 |
247165_at |
AT5G65790
|
ATMYB68, MYB DOMAIN PROTEIN 68, MYB68, myb domain protein 68 |
-0.297 |
-1.143 |
| 55 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.287 |
-0.603 |
| 56 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.268 |
-0.475 |
| 57 |
248801_at |
AT5G47370
|
HAT2 |
-0.249 |
0.153 |
| 58 |
246933_at |
AT5G25160
|
ZFP3, zinc finger protein 3 |
-0.247 |
-0.176 |
| 59 |
251342_at |
AT3G60690
|
SAUR59, SMALL AUXIN UPREGULATED RNA 59 |
-0.237 |
-0.408 |
| 60 |
246566_at |
AT5G14940
|
[Major facilitator superfamily protein] |
-0.228 |
-0.637 |
| 61 |
245906_at |
AT5G11070
|
unknown |
-0.227 |
0.096 |
| 62 |
259596_at |
AT1G28130
|
GH3.17 |
-0.225 |
0.316 |
| 63 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.202 |
-0.662 |
| 64 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.196 |
0.443 |
| 65 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
-0.188 |
-1.059 |
| 66 |
265872_at |
AT2G01670
|
atnudt17, nudix hydrolase homolog 17, NUDT17, nudix hydrolase homolog 17 |
-0.188 |
-0.403 |
| 67 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.187 |
-0.722 |
| 68 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.180 |
0.529 |
| 69 |
253253_at |
AT4G34750
|
SAUR49, SMALL AUXIN UPREGULATED RNA 49 |
-0.180 |
0.749 |
| 70 |
256185_at |
AT1G51700
|
ADOF1, DOF zinc finger protein 1, DOF1, DOF zinc finger protein 1 |
-0.173 |
-0.115 |
| 71 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.169 |
-0.768 |
| 72 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.166 |
-0.169 |
| 73 |
247543_at |
AT5G61600
|
ERF104, ethylene response factor 104 |
-0.162 |
-0.166 |
| 74 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.156 |
-0.409 |
| 75 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.156 |
-0.288 |
| 76 |
257076_at |
AT3G19680
|
unknown |
-0.153 |
0.091 |
| 77 |
250620_at |
AT5G07190
|
ATS3, seed gene 3 |
-0.151 |
-0.797 |
| 78 |
246289_at |
AT3G56880
|
[VQ motif-containing protein] |
-0.150 |
1.003 |
| 79 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
-0.145 |
-0.562 |
| 80 |
246600_at |
AT5G14930
|
SAG101, senescence-associated gene 101 |
-0.141 |
0.011 |
| 81 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
-0.138 |
-0.406 |
| 82 |
250002_at |
AT5G18690
|
AGP25, arabinogalactan protein 25, ATAGP25, ARABIDOPSIS THALIANA ARABINOGALACTAN PROTEINS 25 |
-0.136 |
0.304 |
| 83 |
262166_at |
AT1G74840
|
[Homeodomain-like superfamily protein] |
-0.134 |
0.039 |
| 84 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.134 |
0.547 |
| 85 |
256408_at |
AT1G66610
|
[TRAF-like superfamily protein] |
-0.134 |
-0.259 |
| 86 |
266721_at |
AT2G03220
|
ATFT1, ARABIDOPSIS THALIANA FUCOSYLTRANSFERASE 1, ATFUT1, ARABIDOPSIS THALIANA FUCOSYLTRANSFERASE 1, FT1, fucosyltransferase 1, MUR2, MURUS 2 |
-0.131 |
1.151 |
| 87 |
245831_at |
AT1G48840
|
unknown |
-0.131 |
0.672 |
| 88 |
253217_at |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.127 |
-0.646 |
| 89 |
251533_at |
AT3G58460
|
ATRBL11, ARABIDOPSIS RHOMBOID-LIKE PROTEIN 11, ATRBL15, RHOMBOID-like protein 15, RBL11, rhomboid-like protein 11, RBL15, RHOMBOID-like protein 15 |
-0.127 |
0.764 |
| 90 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.123 |
-0.823 |
| 91 |
248558_at |
AT5G49990
|
[Xanthine/uracil permease family protein] |
-0.122 |
0.143 |
| 92 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
-0.117 |
-0.665 |
| 93 |
265892_at |
AT2G15020
|
unknown |
-0.116 |
0.021 |