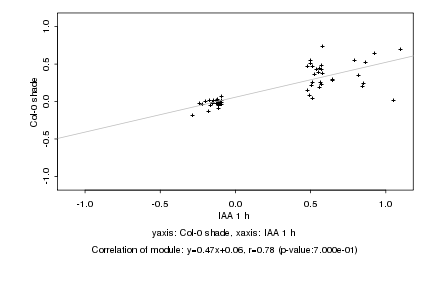
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
IAA 1 h |
Log2 signal ratio
Col-0 shade |
| 1 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
1.095 |
0.701 |
| 2 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
1.051 |
0.016 |
| 3 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.921 |
0.646 |
| 4 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
0.862 |
0.529 |
| 5 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
0.850 |
0.253 |
| 6 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
0.843 |
0.204 |
| 7 |
245076_at |
AT2G23170
|
GH3.3 |
0.816 |
0.360 |
| 8 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.790 |
0.555 |
| 9 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
0.641 |
0.300 |
| 10 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
0.640 |
0.283 |
| 11 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.577 |
0.737 |
| 12 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
0.573 |
0.382 |
| 13 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.571 |
0.237 |
| 14 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.569 |
0.438 |
| 15 |
247925_at |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
0.566 |
0.486 |
| 16 |
252765_at |
AT3G42800
|
unknown |
0.564 |
0.266 |
| 17 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
0.557 |
0.442 |
| 18 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.553 |
0.188 |
| 19 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
0.550 |
0.398 |
| 20 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
0.533 |
0.429 |
| 21 |
248801_at |
AT5G47370
|
HAT2 |
0.523 |
0.363 |
| 22 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
0.512 |
0.050 |
| 23 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
0.508 |
0.470 |
| 24 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.506 |
0.255 |
| 25 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
0.503 |
0.214 |
| 26 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
0.499 |
0.555 |
| 27 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.498 |
0.510 |
| 28 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.487 |
0.084 |
| 29 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
0.473 |
0.153 |
| 30 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
0.473 |
0.469 |
| 31 |
262811_at |
AT1G11700
|
unknown |
-0.291 |
-0.183 |
| 32 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.245 |
-0.019 |
| 33 |
251084_at |
AT5G01520
|
AIRP2, ABA Insensitive RING Protein 2, AtAIRP2 |
-0.220 |
-0.038 |
| 34 |
262166_at |
AT1G74840
|
[Homeodomain-like superfamily protein] |
-0.203 |
0.006 |
| 35 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
-0.180 |
-0.122 |
| 36 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-0.177 |
0.027 |
| 37 |
266479_at |
AT2G31160
|
LSH3, LIGHT SENSITIVE HYPOCOTYLS 3, OBO1, ORGAN BOUNDARY 1 |
-0.169 |
-0.048 |
| 38 |
247397_at |
AT5G62940
|
DOF5.6, DNA BINDING WITH ONE FINGER 5.6, HCA2, HIGH CAMBIAL ACTIVITY2 |
-0.158 |
-0.003 |
| 39 |
259169_at |
AT3G03520
|
NPC3, non-specific phospholipase C3 |
-0.154 |
-0.016 |
| 40 |
246580_at |
AT1G31770
|
ABCG14, ATP-binding cassette G14 |
-0.150 |
0.024 |
| 41 |
252374_at |
AT3G48100
|
ARR5, response regulator 5, ATRR2, ARABIDOPSIS THALIANA RESPONSE REGULATOR 2, IBC6, INDUCED BY CYTOKININ 6, RR5, response regulator 5 |
-0.129 |
0.024 |
| 42 |
248373_at |
AT5G51860
|
AGL72, AGAMOUS-like 72 |
-0.127 |
-0.022 |
| 43 |
254246_at |
AT4G23250
|
CRK17, CYSTEINE-RICH RLK (RECEPTOR-LIKE PROTEIN KINASE) 17, DUF26-21, EMB1290, EMBRYO DEFECTIVE 1290, RKC1, RECEPTOR-LIKE KINASE THALIANACOL-0GENOMICLIBRARY(CLONTECH).THEPOSITIVEPHAGECLONES C-X8-C-X2-C CLASS 1 |
-0.127 |
-0.025 |
| 44 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.124 |
-0.018 |
| 45 |
267347_at |
AT2G39950
|
unknown |
-0.122 |
0.038 |
| 46 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.120 |
0.012 |
| 47 |
245099_at |
AT2G40830
|
RHC1A, RING-H2 finger C1A |
-0.118 |
-0.009 |
| 48 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.118 |
-0.081 |
| 49 |
253022_at |
AT4G38060
|
CCI2, Clavata complex interactor 2 |
-0.116 |
-0.006 |
| 50 |
252025_at |
AT3G52900
|
unknown |
-0.115 |
-0.052 |
| 51 |
253599_at |
AT4G30860
|
ASHR3, ASH1-related 3, SDG4, SET domain group 4 |
-0.109 |
-0.031 |
| 52 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.108 |
-0.019 |
| 53 |
247407_at |
AT5G62900
|
unknown |
-0.107 |
-0.029 |
| 54 |
250232_at |
AT5G13950
|
unknown |
-0.106 |
-0.009 |
| 55 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.102 |
-0.034 |
| 56 |
246997_at |
AT5G67390
|
unknown |
-0.097 |
-0.007 |
| 57 |
249408_at |
AT5G40330
|
ATMYB23, MYB DOMAIN PROTEIN 23, ATMYBRTF, MYB23, myb domain protein 23 |
-0.096 |
0.068 |
| 58 |
263333_at |
AT2G03890
|
ATPI4K GAMMA 7, phosphoinositide 4-kinase gamma 7, PI4K GAMMA 7, phosphoinositide 4-kinase gamma 7, UBDK GAMMA 7, UBIQUITIN-LIKE DOMAIN KINASE GAMMA 7 |
-0.094 |
-0.038 |
| 59 |
256455_at |
AT1G75190
|
unknown |
-0.094 |
0.025 |