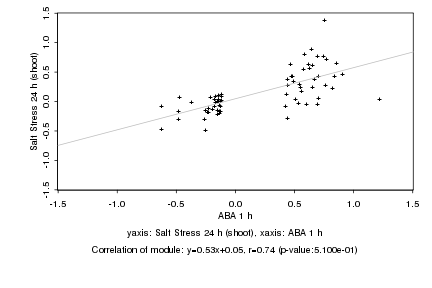
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
ABA 1 h |
Log2 signal ratio
Salt Stress 24 h (shoot) |
| 1 |
258325_at |
AT3G22830
|
AT-HSFA6B, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A6B, HSFA6B, heat shock transcription factor A6B |
1.216 |
0.038 |
| 2 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.905 |
0.469 |
| 3 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
0.854 |
0.661 |
| 4 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.831 |
0.432 |
| 5 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.821 |
0.227 |
| 6 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.768 |
0.717 |
| 7 |
261564_at |
AT1G01720
|
ANAC002, Arabidopsis NAC domain containing protein 2, ATAF1 |
0.756 |
0.279 |
| 8 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
0.749 |
1.393 |
| 9 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.738 |
0.777 |
| 10 |
248203_at |
AT5G54230
|
AtMYB49, myb domain protein 49, MYB49, myb domain protein 49 |
0.696 |
0.057 |
| 11 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.696 |
0.438 |
| 12 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.691 |
-0.048 |
| 13 |
265276_at |
AT2G28400
|
unknown |
0.687 |
0.774 |
| 14 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.666 |
0.383 |
| 15 |
262164_at |
AT1G78070
|
[Transducin/WD40 repeat-like superfamily protein] |
0.649 |
0.241 |
| 16 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.646 |
0.619 |
| 17 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.643 |
0.900 |
| 18 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.621 |
0.563 |
| 19 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.612 |
0.644 |
| 20 |
253851_at |
AT4G28110
|
AtMYB41, myb domain protein 41, MYB41, myb domain protein 41 |
0.598 |
-0.039 |
| 21 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
0.576 |
0.799 |
| 22 |
255723_at |
AT3G29575
|
AFP3, ABI five binding protein 3 |
0.570 |
0.551 |
| 23 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.556 |
0.182 |
| 24 |
251644_at |
AT3G57540
|
[Remorin family protein] |
0.548 |
0.249 |
| 25 |
253994_at |
AT4G26080
|
ABI1, ABA INSENSITIVE 1, AtABI1 |
0.539 |
0.299 |
| 26 |
257274_at |
AT3G14510
|
[Polyprenyl synthetase family protein] |
0.525 |
-0.017 |
| 27 |
248209_at |
AT5G53990
|
[UDP-Glycosyltransferase superfamily protein] |
0.502 |
0.046 |
| 28 |
259231_at |
AT3G11410
|
AHG3, ATPP2CA, ARABIDOPSIS THALIANA PROTEIN PHOSPHATASE 2CA, PP2CA, protein phosphatase 2CA |
0.484 |
0.348 |
| 29 |
252321_at |
AT3G48510
|
unknown |
0.478 |
0.430 |
| 30 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.472 |
0.438 |
| 31 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
0.458 |
0.634 |
| 32 |
263486_at |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
0.439 |
-0.282 |
| 33 |
267538_at |
AT2G41870
|
[Remorin family protein] |
0.435 |
0.283 |
| 34 |
260144_at |
AT1G71960
|
ABCG25, ATP-binding casette G25, ATABCG25, Arabidopsis thaliana ATP-binding cassette G25 |
0.432 |
0.390 |
| 35 |
262452_at |
AT1G11210
|
unknown |
0.424 |
0.122 |
| 36 |
245363_at |
AT4G15120
|
[VQ motif-containing protein] |
0.418 |
-0.068 |
| 37 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.633 |
-0.469 |
| 38 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.630 |
-0.079 |
| 39 |
250777_at |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-0.486 |
-0.162 |
| 40 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.486 |
-0.291 |
| 41 |
247024_at |
AT5G66985
|
unknown |
-0.478 |
0.074 |
| 42 |
260451_at |
AT1G72360
|
AtERF73, ERF73, ethylene response factor 73, HRE1, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 1 |
-0.378 |
-0.006 |
| 43 |
263632_at |
AT2G04795
|
unknown |
-0.270 |
-0.299 |
| 44 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
-0.257 |
-0.480 |
| 45 |
265494_at |
AT2G15680
|
AtCML30, CML30, calmodulin-like 30 |
-0.255 |
-0.148 |
| 46 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
-0.239 |
-0.172 |
| 47 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.236 |
-0.170 |
| 48 |
266803_at |
AT2G28930
|
APK1B, protein kinase 1B, PK1B, protein kinase 1B |
-0.229 |
-0.108 |
| 49 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
-0.219 |
0.074 |
| 50 |
254016_at |
AT4G26150
|
CGA1, cytokinin-responsive gata factor 1, GATA22, GATA TRANSCRIPTION FACTOR 22, GNL, GNC-LIKE |
-0.196 |
-0.121 |
| 51 |
250462_at |
AT5G10020
|
SIRK1, sucrose-induced receptor kinase 1 |
-0.184 |
0.037 |
| 52 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
-0.178 |
-0.074 |
| 53 |
260819_at |
AT1G06850
|
AtbZIP52, basic leucine-zipper 52, bZIP52, basic leucine-zipper 52 |
-0.174 |
0.088 |
| 54 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
-0.171 |
-0.013 |
| 55 |
259523_at |
AT1G12500
|
[Nucleotide-sugar transporter family protein] |
-0.158 |
-0.145 |
| 56 |
256958_at |
AT3G13430
|
[RING/U-box superfamily protein] |
-0.156 |
0.015 |
| 57 |
252272_at |
AT3G49670
|
BAM2, BARELY ANY MERISTEM 2 |
-0.156 |
-0.212 |
| 58 |
246398_at |
AT1G58100
|
TCP8, TCP domain protein 8 |
-0.154 |
0.011 |
| 59 |
246786_at |
AT5G27410
|
[D-aminoacid aminotransferase-like PLP-dependent enzymes superfamily protein] |
-0.150 |
0.110 |
| 60 |
259546_at |
AT1G35350
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.148 |
0.038 |
| 61 |
265944_at |
AT2G19640
|
ASHR2, ASH1-related 2, SDG39, SET DOMAIN PROTEIN 39 |
-0.147 |
0.015 |
| 62 |
266131_at |
AT2G45160
|
ATHAM1, ARABIDOPSIS THALIANA HAIRY MERISTEM 1, HAM1, HAIRY MERISTEM 1, LOM1, LOST MERISTEMS 1 |
-0.144 |
0.035 |
| 63 |
259909_at |
AT1G60870
|
MEE9, maternal effect embryo arrest 9 |
-0.138 |
-0.053 |
| 64 |
250503_at |
AT5G09820
|
[Plastid-lipid associated protein PAP / fibrillin family protein] |
-0.138 |
-0.169 |
| 65 |
247638_at |
AT5G60490
|
AtFLA12, FLA12, FASCICLIN-like arabinogalactan-protein 12 |
-0.135 |
-0.194 |
| 66 |
253252_at |
AT4G34740
|
ASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATASE2, GLN phosphoribosyl pyrophosphate amidotransferase 2, ATPURF2, CIA1, CHLOROPLAST IMPORT APPARATUS 1, DOV1, DIFFERENTIAL DEVELOPMENT OF VASCULAR ASSOCIATED CELLS |
-0.134 |
-0.164 |
| 67 |
256564_at |
AT3G29770
|
ATMES11, ARABIDOPSIS THALIANA METHYL ESTERASE 11, MES11, methyl esterase 11 |
-0.134 |
0.003 |
| 68 |
262649_at |
AT1G14040
|
[EXS (ERD1/XPR1/SYG1) family protein] |
-0.131 |
-0.071 |
| 69 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
-0.125 |
0.032 |
| 70 |
248063_at |
AT5G55550
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.120 |
0.132 |
| 71 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
-0.119 |
0.098 |