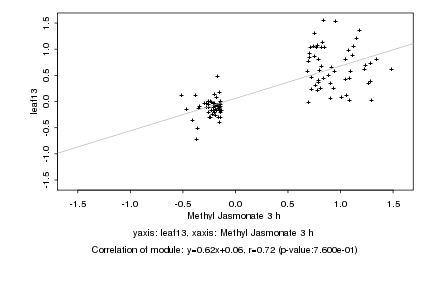
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Methyl Jasmonate 3 h |
Log2 signal ratio
leaf13 |
| 1 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
1.479 |
0.629 |
| 2 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.337 |
0.822 |
| 3 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
1.290 |
0.032 |
| 4 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
1.284 |
0.388 |
| 5 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.276 |
0.740 |
| 6 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
1.258 |
0.354 |
| 7 |
259866_at |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
1.232 |
0.690 |
| 8 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
1.223 |
0.617 |
| 9 |
250292_at |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
1.178 |
1.368 |
| 10 |
264886_at |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
1.143 |
1.220 |
| 11 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
1.120 |
1.067 |
| 12 |
254773_at |
AT4G13410
|
ATCSLA15, CSLA15, CELLULOSE SYNTHASE LIKE A15 |
1.107 |
0.884 |
| 13 |
249071_at |
AT5G44050
|
[MATE efflux family protein] |
1.090 |
0.585 |
| 14 |
259609_at |
AT1G52410
|
AtTSA1, TSA1, TSK-associating protein 1 |
1.079 |
0.451 |
| 15 |
256527_at |
AT1G66100
|
[Plant thionin] |
1.076 |
0.035 |
| 16 |
255527_at |
AT4G02360
|
unknown |
1.069 |
0.988 |
| 17 |
256603_at |
AT3G28270
|
unknown |
1.051 |
0.131 |
| 18 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
1.042 |
0.427 |
| 19 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
1.038 |
0.820 |
| 20 |
254396_at |
AT4G21680
|
AtNPF7.2, NPF7.2, NRT1/ PTR family 7.2, NRT1.8, NITRATE TRANSPORTER 1.8 |
1.001 |
0.083 |
| 21 |
264415_at |
AT1G43160
|
RAP2.6, related to AP2 6 |
0.947 |
1.543 |
| 22 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.940 |
0.590 |
| 23 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.929 |
0.255 |
| 24 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
0.909 |
0.662 |
| 25 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.898 |
0.357 |
| 26 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.897 |
0.068 |
| 27 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.881 |
0.509 |
| 28 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.842 |
1.033 |
| 29 |
266901_at |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
0.833 |
1.567 |
| 30 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.831 |
0.457 |
| 31 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.821 |
1.145 |
| 32 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.817 |
1.046 |
| 33 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.810 |
0.680 |
| 34 |
250455_at |
AT5G09980
|
PROPEP4, elicitor peptide 4 precursor |
0.807 |
0.253 |
| 35 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
0.792 |
0.611 |
| 36 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.790 |
0.377 |
| 37 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
0.789 |
0.810 |
| 38 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.788 |
0.404 |
| 39 |
253684_at |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
0.776 |
0.218 |
| 40 |
260399_at |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
0.775 |
1.081 |
| 41 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.766 |
1.039 |
| 42 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.761 |
0.316 |
| 43 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.746 |
1.306 |
| 44 |
247047_at |
AT5G66650
|
unknown |
0.745 |
0.873 |
| 45 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.736 |
1.066 |
| 46 |
259568_at |
AT1G20490
|
[AMP-dependent synthetase and ligase family protein] |
0.722 |
0.238 |
| 47 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.716 |
0.465 |
| 48 |
266246_at |
AT2G27690
|
CYP94C1, cytochrome P450, family 94, subfamily C, polypeptide 1 |
0.711 |
1.036 |
| 49 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
0.698 |
0.857 |
| 50 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.696 |
0.926 |
| 51 |
262653_at |
AT1G14130
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.693 |
0.777 |
| 52 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.686 |
-0.013 |
| 53 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.679 |
0.577 |
| 54 |
264636_at |
AT1G65490
|
unknown |
-0.517 |
0.125 |
| 55 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.468 |
-0.151 |
| 56 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.418 |
-0.350 |
| 57 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.388 |
0.126 |
| 58 |
266745_at |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.374 |
-0.713 |
| 59 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.363 |
-0.500 |
| 60 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.352 |
-0.125 |
| 61 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.348 |
-0.091 |
| 62 |
259801_at |
AT1G72230
|
[Cupredoxin superfamily protein] |
-0.303 |
-0.024 |
| 63 |
264470_at |
AT1G67110
|
CYP735A2, cytochrome P450, family 735, subfamily A, polypeptide 2 |
-0.281 |
-0.030 |
| 64 |
262388_at |
AT1G49320
|
unknown |
-0.280 |
-0.111 |
| 65 |
267592_at |
AT2G39710
|
[Eukaryotic aspartyl protease family protein] |
-0.265 |
-0.108 |
| 66 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
-0.260 |
-0.203 |
| 67 |
253437_at |
AT4G32460
|
unknown |
-0.259 |
0.006 |
| 68 |
265083_at |
AT1G03820
|
unknown |
-0.258 |
-0.045 |
| 69 |
248273_at |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.249 |
-0.295 |
| 70 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.242 |
-0.301 |
| 71 |
252037_at |
AT3G51920
|
ATCML9, CAM9, calmodulin 9, CML9, CALMODULIN LIKE PROTEIN 9 |
-0.238 |
0.004 |
| 72 |
266447_at |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.236 |
-0.160 |
| 73 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.236 |
-0.006 |
| 74 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.226 |
-0.247 |
| 75 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.211 |
-0.171 |
| 76 |
249885_at |
AT5G22940
|
F8H, FRA8 homolog |
-0.210 |
-0.036 |
| 77 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.209 |
-0.112 |
| 78 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.207 |
-0.194 |
| 79 |
255926_at |
AT1G22190
|
RAP2.4, related to AP2 4 |
-0.207 |
-0.075 |
| 80 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.206 |
0.142 |
| 81 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
-0.200 |
-0.028 |
| 82 |
258409_at |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.199 |
-0.254 |
| 83 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.196 |
-0.136 |
| 84 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.195 |
-0.076 |
| 85 |
252126_at |
AT3G50950
|
ZAR1, HOPZ-ACTIVATED RESISTANCE 1 |
-0.185 |
0.093 |
| 86 |
256525_at |
AT1G66180
|
[Eukaryotic aspartyl protease family protein] |
-0.182 |
-0.066 |
| 87 |
246387_at |
AT1G77400
|
unknown |
-0.181 |
-0.170 |
| 88 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.180 |
-0.063 |
| 89 |
248611_at |
AT5G49520
|
ATWRKY48, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 48, WRKY48, WRKY DNA-binding protein 48 |
-0.179 |
0.493 |
| 90 |
247049_at |
AT5G66440
|
unknown |
-0.172 |
-0.068 |
| 91 |
266007_at |
AT2G37380
|
MAKR3, MEMBRANE-ASSOCIATED KINASE REGULATOR 3 |
-0.170 |
-0.302 |
| 92 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.166 |
-0.128 |
| 93 |
248003_at |
AT5G56220
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.164 |
-0.087 |
| 94 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.161 |
-0.167 |
| 95 |
252943_at |
AT4G39330
|
ATCAD9, CAD9, cinnamyl alcohol dehydrogenase 9 |
-0.156 |
-0.033 |
| 96 |
256336_at |
AT1G72030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.156 |
-0.101 |
| 97 |
253185_at |
AT4G35240
|
unknown |
-0.155 |
0.189 |
| 98 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.152 |
-0.394 |
| 99 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.148 |
-0.171 |
| 100 |
261598_at |
AT1G49750
|
[Leucine-rich repeat (LRR) family protein] |
-0.148 |
-0.077 |
| 101 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.148 |
-0.293 |
| 102 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.146 |
-0.038 |
| 103 |
252343_at |
AT3G48610
|
NPC6, non-specific phospholipase C6 |
-0.144 |
0.015 |
| 104 |
264900_at |
AT1G23080
|
ATPIN7, ARABIDOPSIS PIN-FORMED 7, PIN7, PIN-FORMED 7 |
-0.143 |
-0.208 |
| 105 |
262039_at |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
-0.142 |
-0.164 |