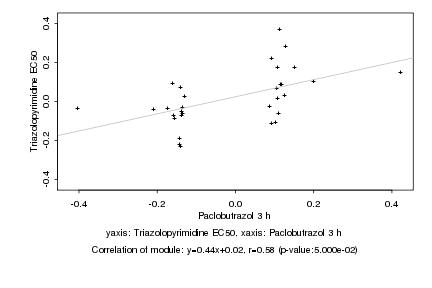
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Paclobutrazol 3 h |
Log2 signal ratio
Triazolopyrimidine EC50 |
| 1 |
256598_at |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
0.421 |
0.154 |
| 2 |
256700_at |
AT3G52260
|
[Pseudouridine synthase family protein] |
0.198 |
0.105 |
| 3 |
260273_at |
AT1G80550
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.149 |
0.179 |
| 4 |
248136_at |
AT5G54910
|
[DEA(D/H)-box RNA helicase family protein] |
0.127 |
0.287 |
| 5 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
0.125 |
0.035 |
| 6 |
262942_at |
AT1G79450
|
ALIS5, ALA-interacting subunit 5 |
0.116 |
0.092 |
| 7 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.113 |
0.090 |
| 8 |
250555_at |
AT5G07950
|
unknown |
0.112 |
0.372 |
| 9 |
253985_at |
AT4G26220
|
CCoAOMT7, caffeoyl coenzyme A ester O-methyltransferase 7 |
0.109 |
-0.060 |
| 10 |
262774_at |
AT1G13230
|
[Leucine-rich repeat (LRR) family protein] |
0.106 |
0.018 |
| 11 |
267490_at |
AT2G19130
|
[S-locus lectin protein kinase family protein] |
0.105 |
0.175 |
| 12 |
253029_at |
AT4G38170
|
FRS9, FAR1-related sequence 9 |
0.104 |
0.070 |
| 13 |
252418_at |
AT3G47490
|
[HNH endonuclease] |
0.101 |
-0.105 |
| 14 |
246674_at |
AT5G30490
|
unknown |
0.092 |
0.221 |
| 15 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
0.090 |
-0.111 |
| 16 |
255824_at |
AT2G40530
|
unknown |
0.085 |
-0.022 |
| 17 |
251125_at |
AT5G01060
|
BSK10, brassinosteroid-signaling kinase 10 |
-0.406 |
-0.034 |
| 18 |
263733_at |
AT1G60020
|
[copia-like retrotransposon family, has a 0. P-value blast match to GB:AAC02672 polyprotein (Ty1_Copia-element) (Arabidopsis arenosa)] |
-0.210 |
-0.036 |
| 19 |
251268_at |
AT3G62350
|
unknown |
-0.176 |
-0.034 |
| 20 |
249222_at |
AT5G42450
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.161 |
0.094 |
| 21 |
252286_at |
AT3G49070
|
unknown |
-0.159 |
-0.069 |
| 22 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
-0.157 |
-0.087 |
| 23 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.145 |
-0.186 |
| 24 |
263443_at |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-0.145 |
-0.217 |
| 25 |
267634_at |
AT2G42100
|
[Actin-like ATPase superfamily protein] |
-0.141 |
0.072 |
| 26 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.141 |
-0.228 |
| 27 |
254906_at |
AT4G11180
|
DP1, dirigent protein 1 |
-0.139 |
-0.050 |
| 28 |
265211_at |
AT2G36640
|
ATECP63, embryonic cell protein 63, ECP63, embryonic cell protein 63 |
-0.139 |
-0.071 |
| 29 |
253710_at |
AT4G29230
|
anac075, NAC domain containing protein 75, NAC075, NAC domain containing protein 75 |
-0.137 |
-0.026 |
| 30 |
256449_at |
AT1G33460
|
[Mutator-like transposase family, has a 3.6e-40 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-0.136 |
-0.060 |
| 31 |
261516_at |
AT1G71750
|
HGPT, Hypoxanthine-guanine phosphoribosyltransferase |
-0.131 |
0.028 |