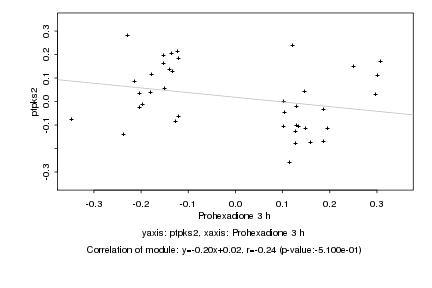
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Prohexadione 3 h |
Log2 signal ratio
ptpks2 |
| 1 |
250669_at |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
0.307 |
0.172 |
| 2 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
0.299 |
0.115 |
| 3 |
246620_at |
AT5G36220
|
CYP81D1, cytochrome P450, family 81, subfamily D, polypeptide 1, CYP91A1, CYTOCHROME P450 91A1 |
0.295 |
0.034 |
| 4 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.249 |
0.150 |
| 5 |
261026_at |
AT1G01240
|
unknown |
0.195 |
-0.113 |
| 6 |
258395_at |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
0.186 |
-0.166 |
| 7 |
254922_at |
AT4G11370
|
RHA1A, RING-H2 finger A1A |
0.185 |
-0.030 |
| 8 |
249879_at |
AT5G23170
|
[Protein kinase superfamily protein] |
0.158 |
-0.172 |
| 9 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.147 |
-0.114 |
| 10 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
0.145 |
0.045 |
| 11 |
246592_at |
AT5G14890
|
[NHL domain-containing protein] |
0.132 |
-0.104 |
| 12 |
255824_at |
AT2G40530
|
unknown |
0.128 |
-0.101 |
| 13 |
248741_at |
AT5G48170
|
SLY2, SLEEPY2, SNE, SNEEZY |
0.128 |
-0.017 |
| 14 |
267438_at |
AT2G19050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.127 |
-0.178 |
| 15 |
259073_at |
AT3G02290
|
[RING/U-box superfamily protein] |
0.126 |
-0.124 |
| 16 |
245816_at |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
0.119 |
0.240 |
| 17 |
250158_at |
AT5G15190
|
unknown |
0.114 |
-0.257 |
| 18 |
244906_at |
ATMG00690
|
ORF240A |
0.103 |
-0.043 |
| 19 |
256667_at |
AT3G32180
|
unknown |
0.101 |
-0.106 |
| 20 |
259451_at |
AT1G13890
|
ATSNAP30, SNAP30, soluble N-ethylmaleimide-sensitive factor adaptor protein 30 |
0.101 |
0.003 |
| 21 |
263836_at |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-0.349 |
-0.074 |
| 22 |
248709_at |
AT5G48470
|
PRDA1, PEP-Related Development Arrested 1 |
-0.239 |
-0.138 |
| 23 |
253917_at |
AT4G27380
|
unknown |
-0.230 |
0.281 |
| 24 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.215 |
0.089 |
| 25 |
247057_at |
AT5G66770
|
[GRAS family transcription factor] |
-0.204 |
-0.022 |
| 26 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.204 |
0.037 |
| 27 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.199 |
-0.011 |
| 28 |
258747_at |
AT3G05810
|
unknown |
-0.182 |
0.042 |
| 29 |
258522_at |
AT3G06660
|
[PAPA-1-like family protein / zinc finger (HIT type) family protein] |
-0.178 |
0.118 |
| 30 |
265819_at |
AT2G17972
|
unknown |
-0.154 |
0.198 |
| 31 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
-0.153 |
0.165 |
| 32 |
250452_at |
AT5G10630
|
[Translation elongation factor EF1A/initiation factor IF2gamma family protein] |
-0.151 |
0.057 |
| 33 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
-0.140 |
0.140 |
| 34 |
259392_at |
AT1G06380
|
[Ribosomal protein L1p/L10e family] |
-0.137 |
0.206 |
| 35 |
249144_at |
AT5G43270
|
SPL2, squamosa promoter binding protein-like 2 |
-0.134 |
0.128 |
| 36 |
256284_at |
AT3G12530
|
PSF2 |
-0.128 |
-0.082 |
| 37 |
247032_at |
AT5G67240
|
SDN3, small RNA degrading nuclease 3 |
-0.123 |
0.217 |
| 38 |
248549_at |
AT5G50310
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.122 |
0.186 |
| 39 |
251291_at |
AT3G61900
|
SAUR33, SMALL AUXIN UPREGULATED RNA 33 |
-0.122 |
-0.063 |