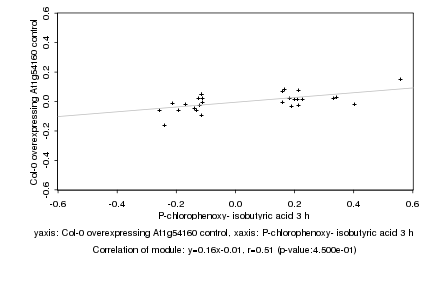
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
P-chlorophenoxy- isobutyric acid 3 h |
Log2 signal ratio
Col-0 overexpressing At1g54160 control |
| 1 |
251677_at |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
0.557 |
0.155 |
| 2 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.401 |
-0.016 |
| 3 |
257823_at |
AT3G25190
|
[Vacuolar iron transporter (VIT) family protein] |
0.339 |
0.032 |
| 4 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.330 |
0.025 |
| 5 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.226 |
0.016 |
| 6 |
266296_at |
AT2G29420
|
ATGSTU7, glutathione S-transferase tau 7, GST25, GLUTATHIONE S-TRANSFERASE 25, GSTU7, glutathione S-transferase tau 7 |
0.213 |
0.076 |
| 7 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.213 |
-0.023 |
| 8 |
259228_at |
AT3G07720
|
[Galactose oxidase/kelch repeat superfamily protein] |
0.208 |
0.018 |
| 9 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
0.198 |
0.020 |
| 10 |
248750_at |
AT5G47530
|
[Auxin-responsive family protein] |
0.188 |
-0.029 |
| 11 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.182 |
0.026 |
| 12 |
255726_at |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
0.165 |
0.085 |
| 13 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
0.158 |
0.071 |
| 14 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.158 |
-0.002 |
| 15 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.258 |
-0.055 |
| 16 |
249097_at |
AT5G43520
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.240 |
-0.161 |
| 17 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
-0.214 |
-0.011 |
| 18 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.193 |
-0.057 |
| 19 |
255936_at |
AT1G12680
|
PEPKR2, phosphoenolpyruvate carboxylase-related kinase 2 |
-0.170 |
-0.017 |
| 20 |
253732_at |
AT4G29140
|
ADS1, ACTIVATED DISEASE SUSCEPTIBILITY 1 |
-0.140 |
-0.045 |
| 21 |
251838_at |
AT3G54940
|
[Papain family cysteine protease] |
-0.135 |
-0.059 |
| 22 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.128 |
0.023 |
| 23 |
260178_at |
AT1G70720
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.124 |
-0.021 |
| 24 |
259528_at |
AT1G12330
|
unknown |
-0.117 |
-0.090 |
| 25 |
246788_at |
AT5G27580
|
AGL89, AGAMOUS-like 89 |
-0.116 |
0.050 |
| 26 |
253149_at |
AT4G35650
|
IDH-III, isocitrate dehydrogenase III |
-0.115 |
0.048 |
| 27 |
248145_at |
AT5G54880
|
[DTW domain-containing protein] |
-0.114 |
-0.006 |
| 28 |
256887_at |
AT3G15150
|
ATMMS21, A. THALIANA METHYL METHANE SULFONATE SENSITIVITY 21, HPY2, HIGH PLOIDY2, MMS21, METHYL METHANE SULFONATE SENSITIVITY 2 |
-0.112 |
0.027 |