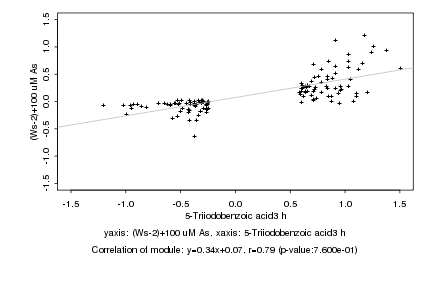
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
5-Triiodobenzoic acid3 h |
Log2 signal ratio
(Ws-2)+100 uM As |
| 1 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
1.502 |
0.621 |
| 2 |
253046_at |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
1.372 |
0.950 |
| 3 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
1.256 |
1.020 |
| 4 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
1.236 |
0.905 |
| 5 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
1.197 |
0.177 |
| 6 |
262148_at |
AT1G52560
|
[HSP20-like chaperones superfamily protein] |
1.174 |
1.217 |
| 7 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
1.150 |
0.708 |
| 8 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
1.117 |
0.603 |
| 9 |
258277_at |
AT3G26830
|
CYP71B15, PAD3, PHYTOALEXIN DEFICIENT 3 |
1.100 |
0.160 |
| 10 |
263182_at |
AT1G05575
|
unknown |
1.096 |
0.100 |
| 11 |
264648_at |
AT1G09080
|
BIP3, binding protein 3 |
1.071 |
0.011 |
| 12 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
1.048 |
0.415 |
| 13 |
266294_at |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
1.031 |
0.637 |
| 14 |
265499_at |
AT2G15480
|
UGT73B5, UDP-glucosyl transferase 73B5 |
1.025 |
0.280 |
| 15 |
253044_at |
AT4G37290
|
unknown |
1.025 |
0.743 |
| 16 |
251824_at |
AT3G55090
|
ABCG16, ATP-binding cassette G16 |
1.023 |
0.871 |
| 17 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.959 |
0.237 |
| 18 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.957 |
0.206 |
| 19 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.956 |
0.285 |
| 20 |
262124_at |
AT1G59660
|
[Nucleoporin autopeptidase] |
0.948 |
-0.020 |
| 21 |
260706_at |
AT1G32350
|
AOX1D, alternative oxidase 1D |
0.937 |
0.151 |
| 22 |
261838_at |
AT1G16030
|
Hsp70b, heat shock protein 70B |
0.911 |
0.643 |
| 23 |
260978_at |
AT1G53540
|
[HSP20-like chaperones superfamily protein] |
0.910 |
1.134 |
| 24 |
245076_at |
AT2G23170
|
GH3.3 |
0.909 |
0.514 |
| 25 |
266290_at |
AT2G29490
|
ATGSTU1, glutathione S-transferase TAU 1, GST19, GLUTATHIONE S-TRANSFERASE 19, GSTU1, glutathione S-transferase TAU 1 |
0.905 |
0.250 |
| 26 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.877 |
0.422 |
| 27 |
261763_at |
AT1G15520
|
ABCG40, ATP-binding cassette G40, ATABCG40, Arabidopsis thaliana ATP-binding cassette G40, ATPDR12, PLEIOTROPIC DRUG RESISTANCE 12, PDR12, pleiotropic drug resistance 12 |
0.874 |
0.096 |
| 28 |
261545_at |
AT1G63530
|
unknown |
0.868 |
0.003 |
| 29 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
0.847 |
0.749 |
| 30 |
261892_at |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
0.844 |
0.101 |
| 31 |
253796_at |
AT4G28460
|
unknown |
0.837 |
0.253 |
| 32 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.836 |
0.418 |
| 33 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.834 |
0.464 |
| 34 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.830 |
0.288 |
| 35 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
0.778 |
0.362 |
| 36 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
0.776 |
0.595 |
| 37 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.776 |
0.174 |
| 38 |
245082_at |
AT2G23270
|
unknown |
0.752 |
0.476 |
| 39 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.733 |
0.071 |
| 40 |
257536_at |
AT3G02800
|
AtPFA-DSP3, PFA-DSP3, plant and fungi atypical dual-specificity phosphatase 3 |
0.726 |
0.264 |
| 41 |
246858_at |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.721 |
0.233 |
| 42 |
250770_at |
AT5G05390
|
LAC12, laccase 12 |
0.715 |
0.452 |
| 43 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.710 |
0.684 |
| 44 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.709 |
0.190 |
| 45 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
0.708 |
0.025 |
| 46 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.705 |
0.053 |
| 47 |
249942_at |
AT5G22300
|
AtNIT4, NITRILASE 4, NIT4, nitrilase 4 |
0.690 |
0.115 |
| 48 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.689 |
0.379 |
| 49 |
250252_at |
AT5G13750
|
ZIFL1, zinc induced facilitator-like 1 |
0.670 |
0.283 |
| 50 |
259979_at |
AT1G76600
|
unknown |
0.655 |
0.296 |
| 51 |
256762_at |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
0.650 |
0.306 |
| 52 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.649 |
0.184 |
| 53 |
245352_at |
AT4G15490
|
UGT84A3 |
0.644 |
0.266 |
| 54 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.637 |
0.293 |
| 55 |
245250_at |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
0.633 |
0.198 |
| 56 |
246067_at |
AT5G19410
|
ABCG23, ATP-binding cassette G23 |
0.632 |
0.182 |
| 57 |
267008_at |
AT2G39350
|
ABCG1, ATP-binding cassette G1 |
0.617 |
0.265 |
| 58 |
260643_at |
AT1G53270
|
ABCG10, ATP-binding cassette G10 |
0.614 |
0.273 |
| 59 |
254926_at |
AT4G11280
|
ACS6, 1-aminocyclopropane-1-carboxylic acid (acc) synthase 6, ATACS6 |
0.614 |
0.100 |
| 60 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
0.611 |
0.249 |
| 61 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.610 |
0.302 |
| 62 |
250550_at |
AT5G07870
|
[HXXXD-type acyl-transferase family protein] |
0.599 |
0.248 |
| 63 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.598 |
0.184 |
| 64 |
246203_at |
AT4G36610
|
[alpha/beta-Hydrolases superfamily protein] |
0.597 |
0.344 |
| 65 |
258100_at |
AT3G23550
|
[MATE efflux family protein] |
0.596 |
-0.009 |
| 66 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.590 |
0.132 |
| 67 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.584 |
0.167 |
| 68 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-1.209 |
-0.056 |
| 69 |
254772_at |
AT4G13390
|
EXT12, extensin 12 |
-1.030 |
-0.068 |
| 70 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-1.001 |
-0.221 |
| 71 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.966 |
-0.061 |
| 72 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.951 |
-0.111 |
| 73 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.936 |
-0.054 |
| 74 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
-0.894 |
-0.042 |
| 75 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.861 |
-0.077 |
| 76 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.817 |
-0.095 |
| 77 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.704 |
-0.032 |
| 78 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
-0.649 |
-0.029 |
| 79 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
-0.623 |
-0.048 |
| 80 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.595 |
-0.039 |
| 81 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.593 |
-0.067 |
| 82 |
264404_at |
AT2G25160
|
CYP82F1, cytochrome P450, family 82, subfamily F, polypeptide 1 |
-0.580 |
-0.300 |
| 83 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.558 |
-0.019 |
| 84 |
250778_at |
AT5G05500
|
MOP10 |
-0.547 |
-0.031 |
| 85 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.536 |
-0.259 |
| 86 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.530 |
0.027 |
| 87 |
247059_at |
AT5G66690
|
UGT72E2 |
-0.521 |
-0.041 |
| 88 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
-0.512 |
-0.026 |
| 89 |
253483_at |
AT4G31910
|
BAT1, BR-related AcylTransferase1, PIZ, PIZZA |
-0.504 |
-0.181 |
| 90 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.500 |
0.028 |
| 91 |
266356_at |
AT2G32300
|
UCC1, uclacyanin 1 |
-0.484 |
-0.120 |
| 92 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.448 |
-0.029 |
| 93 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
-0.437 |
-0.138 |
| 94 |
266336_at |
AT2G32270
|
ZIP3, zinc transporter 3 precursor |
-0.434 |
-0.196 |
| 95 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.428 |
0.034 |
| 96 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.427 |
-0.342 |
| 97 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.422 |
-0.157 |
| 98 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
-0.419 |
-0.040 |
| 99 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.410 |
-0.017 |
| 100 |
253667_at |
AT4G30170
|
[Peroxidase family protein] |
-0.380 |
-0.054 |
| 101 |
246142_at |
AT5G19970
|
unknown |
-0.379 |
-0.628 |
| 102 |
251405_at |
AT3G60330
|
AHA7, H(+)-ATPase 7, HA7, H(+)-ATPase 7 |
-0.374 |
-0.022 |
| 103 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.374 |
0.015 |
| 104 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.374 |
-0.069 |
| 105 |
249227_at |
AT5G42180
|
PER64, peroxidase 64 |
-0.365 |
-0.083 |
| 106 |
259786_at |
AT1G29660
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.363 |
-0.039 |
| 107 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.360 |
-0.345 |
| 108 |
258209_at |
AT3G14060
|
unknown |
-0.345 |
-0.244 |
| 109 |
248178_at |
AT5G54370
|
[Late embryogenesis abundant (LEA) protein-related] |
-0.340 |
0.021 |
| 110 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.332 |
0.000 |
| 111 |
245567_at |
AT4G14630
|
GLP9, germin-like protein 9 |
-0.325 |
-0.170 |
| 112 |
251811_at |
AT3G54990
|
SMZ, SCHLAFMUTZE |
-0.323 |
-0.169 |
| 113 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.314 |
-0.016 |
| 114 |
250099_at |
AT5G17300
|
RVE1, REVEILLE 1 |
-0.314 |
0.037 |
| 115 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
-0.312 |
-0.034 |
| 116 |
252046_at |
AT3G52460
|
[hydroxyproline-rich glycoprotein family protein] |
-0.304 |
0.032 |
| 117 |
263552_x_at |
AT2G24980
|
EXT6, extensin 6 |
-0.303 |
0.011 |
| 118 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
-0.299 |
-0.011 |
| 119 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.298 |
-0.123 |
| 120 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.294 |
0.002 |
| 121 |
253732_at |
AT4G29140
|
ADS1, ACTIVATED DISEASE SUSCEPTIBILITY 1 |
-0.272 |
-0.141 |
| 122 |
260230_at |
AT1G74500
|
ATBS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, bHLH135, basic helix-loop-helix protein 135, BS1, activation-tagged BRI1(brassinosteroid-insensitive 1)-suppressor 1, PRE3, PACLOBUTRAZOL RESISTANCE 3, TMO7, TARGET OF MONOPTEROS 7 |
-0.272 |
-0.043 |
| 123 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
-0.272 |
-0.044 |
| 124 |
250438_at |
AT5G10580
|
unknown |
-0.269 |
-0.189 |
| 125 |
246997_at |
AT5G67390
|
unknown |
-0.268 |
-0.070 |
| 126 |
257852_at |
AT3G12950
|
[Trypsin family protein] |
-0.267 |
-0.124 |
| 127 |
251174_at |
AT3G63200
|
PLA IIIB, PLP9, PATATIN-like protein 9 |
-0.262 |
-0.049 |
| 128 |
250682_x_at |
AT5G06630
|
[proline-rich extensin-like family protein] |
-0.260 |
-0.000 |
| 129 |
249151_at |
AT5G43360
|
ATPT4, PHT1;3, phosphate transporter 1;3, PHT3, PHOSPHATE TRANSPORTER 3 |
-0.260 |
0.000 |
| 130 |
248150_at |
AT5G54670
|
ATK3, kinesin 3, KATC, KINESIN-LIKE PROTEIN IN ARABIDOPSIS THALIANA C |
-0.257 |
-0.020 |
| 131 |
260077_at |
AT1G73620
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.255 |
-0.133 |
| 132 |
256283_at |
AT3G12540
|
unknown |
-0.254 |
0.003 |
| 133 |
267077_at |
AT2G40970
|
MYBC1 |
-0.250 |
-0.110 |