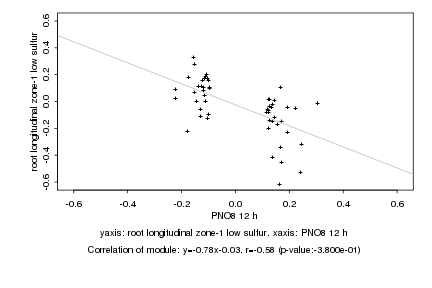
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
PNO8 12 h |
Log2 signal ratio
root longitudinal zone-1 low sulfur |
| 1 |
253161_at |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
0.302 |
-0.014 |
| 2 |
260287_at |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
0.242 |
-0.314 |
| 3 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.239 |
-0.523 |
| 4 |
266625_at |
AT2G35380
|
[Peroxidase superfamily protein] |
0.220 |
-0.049 |
| 5 |
253061_at |
AT4G37610
|
BT5, BTB and TAZ domain protein 5 |
0.193 |
-0.038 |
| 6 |
258402_at |
AT3G15450
|
[Aluminium induced protein with YGL and LRDR motifs] |
0.192 |
-0.224 |
| 7 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
0.170 |
-0.145 |
| 8 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.169 |
-0.451 |
| 9 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
0.166 |
-0.340 |
| 10 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.165 |
0.105 |
| 11 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.160 |
-0.611 |
| 12 |
246034_at |
AT5G08350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
0.156 |
-0.168 |
| 13 |
262674_at |
AT1G75910
|
EXL4, extracellular lipase 4 |
0.144 |
0.008 |
| 14 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
0.143 |
-0.114 |
| 15 |
266504_at |
AT2G47820
|
unknown |
0.137 |
-0.145 |
| 16 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.136 |
-0.416 |
| 17 |
263118_at |
AT1G03090
|
MCCA |
0.135 |
-0.020 |
| 18 |
246697_at |
AT5G29210
|
unknown |
0.132 |
-0.043 |
| 19 |
258012_at |
AT3G19310
|
[PLC-like phosphodiesterases superfamily protein] |
0.126 |
-0.037 |
| 20 |
266337_at |
AT2G32390
|
ATGLR3.5, glutamate receptor 3.5, GLR3.5, glutamate receptor 3.5, GLR6 |
0.125 |
0.020 |
| 21 |
253249_at |
AT4G34680
|
GATA3, GATA transcription factor 3 |
0.125 |
-0.137 |
| 22 |
256793_at |
AT3G22160
|
JAV1, jasmonate-associated VQ motif gene 1 |
0.122 |
-0.197 |
| 23 |
257872_at |
AT3G28360
|
ABCB16, ATP-binding cassette B16, PGP16, P-glycoprotein 16 |
0.121 |
0.017 |
| 24 |
256566_at |
AT3G19530
|
unknown |
0.120 |
-0.062 |
| 25 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.119 |
-0.081 |
| 26 |
265524_at |
AT2G06180
|
[pseudogene] |
0.116 |
-0.056 |
| 27 |
246182_at |
AT5G20870
|
[O-Glycosyl hydrolases family 17 protein] |
0.114 |
-0.076 |
| 28 |
251371_at |
AT3G60360
|
EDA14, EMBRYO SAC DEVELOPMENT ARREST 14, UTP11, U3 SMALL NUCLEOLAR RNA-ASSOCIATED PROTEIN 11 |
-0.224 |
0.028 |
| 29 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
-0.223 |
0.096 |
| 30 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.181 |
-0.216 |
| 31 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
-0.176 |
0.186 |
| 32 |
246461_at |
AT5G16930
|
[AAA-type ATPase family protein] |
-0.158 |
0.335 |
| 33 |
256676_at |
AT3G52180
|
ATPTPKIS1, ATSEX4, DSP4, DUAL-SPECIFICITY PROTEIN PHOSPHATASE 4, SEX4, STARCH-EXCESS 4 |
-0.153 |
0.069 |
| 34 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.152 |
0.282 |
| 35 |
259139_at |
AT3G10240
|
[F-box and associated interaction domains-containing protein] |
-0.146 |
0.006 |
| 36 |
248075_at |
AT5G55740
|
CRR21, chlororespiratory reduction 21 |
-0.138 |
0.113 |
| 37 |
266222_at |
AT2G28780
|
unknown |
-0.132 |
-0.111 |
| 38 |
263188_at |
AT1G36095
|
[DNA binding] |
-0.130 |
-0.058 |
| 39 |
267399_at |
AT2G44195
|
[CBF1-interacting co-repressor CIR, N-terminal] |
-0.128 |
0.117 |
| 40 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
-0.124 |
0.161 |
| 41 |
256065_at |
AT1G07070
|
[Ribosomal protein L35Ae family protein] |
-0.121 |
0.105 |
| 42 |
253607_at |
AT4G30330
|
[Small nuclear ribonucleoprotein family protein] |
-0.119 |
0.087 |
| 43 |
257018_at |
AT3G19630
|
[Radical SAM superfamily protein] |
-0.116 |
0.050 |
| 44 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
-0.116 |
0.177 |
| 45 |
255501_at |
AT4G02400
|
[U3 ribonucleoprotein (Utp) family protein] |
-0.114 |
0.189 |
| 46 |
248984_at |
AT5G45140
|
NRPC2, nuclear RNA polymerase C2 |
-0.113 |
0.007 |
| 47 |
248471_at |
AT5G50840
|
unknown |
-0.110 |
0.202 |
| 48 |
247098_at |
AT5G66470
|
[RNA binding] |
-0.107 |
0.174 |
| 49 |
260220_at |
AT1G74650
|
ATMYB31, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 31, ATY13, MYB31, myb domain protein 31 |
-0.106 |
-0.123 |
| 50 |
255586_at |
AT4G01560
|
MEE49, maternal effect embryo arrest 49 |
-0.102 |
0.164 |
| 51 |
265660_at |
AT2G25470
|
AtRLP21, receptor like protein 21, RLP21, receptor like protein 21 |
-0.100 |
-0.094 |
| 52 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.099 |
0.099 |
| 53 |
249554_at |
AT5G38290
|
[Peptidyl-tRNA hydrolase family protein] |
-0.097 |
0.106 |