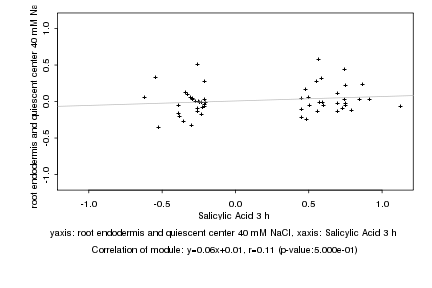
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Salicylic Acid 3 h |
Log2 signal ratio
root endodermis and quiescent center 40 mM NaCl |
| 1 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
1.123 |
-0.066 |
| 2 |
249295_at |
AT5G41280
|
[Receptor-like protein kinase-related family protein] |
0.910 |
0.037 |
| 3 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.864 |
0.235 |
| 4 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.846 |
0.040 |
| 5 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
0.786 |
-0.111 |
| 6 |
265501_at |
AT2G15490
|
UGT73B4, UDP-glycosyltransferase 73B4 |
0.748 |
0.230 |
| 7 |
254093_at |
AT4G25110
|
AtMC2, metacaspase 2, AtMCP1c, metacaspase 1c, MC2, metacaspase 2, MCP1c, metacaspase 1c |
0.745 |
-0.026 |
| 8 |
265698_at |
AT2G32160
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.744 |
-0.049 |
| 9 |
267573_at |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.741 |
0.031 |
| 10 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.740 |
0.446 |
| 11 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.728 |
-0.088 |
| 12 |
253181_at |
AT4G35180
|
LHT7, LYS/HIS transporter 7 |
0.692 |
0.116 |
| 13 |
259251_at |
AT3G07600
|
[Heavy metal transport/detoxification superfamily protein ] |
0.691 |
-0.015 |
| 14 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
0.691 |
-0.123 |
| 15 |
259260_at |
AT3G11370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.600 |
-0.048 |
| 16 |
259454_at |
AT1G44050
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.589 |
-0.001 |
| 17 |
259213_at |
AT3G09010
|
[Protein kinase superfamily protein] |
0.581 |
0.324 |
| 18 |
259385_at |
AT1G13470
|
unknown |
0.570 |
-0.008 |
| 19 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.562 |
0.576 |
| 20 |
254629_at |
AT4G18425
|
unknown |
0.553 |
-0.130 |
| 21 |
262381_at |
AT1G72900
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
0.548 |
0.278 |
| 22 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
0.500 |
-0.050 |
| 23 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.496 |
0.059 |
| 24 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
0.478 |
-0.239 |
| 25 |
267496_at |
AT2G30550
|
DALL3, DAD1-Like Lipase 3 |
0.471 |
0.170 |
| 26 |
263005_at |
AT1G54540
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.449 |
-0.106 |
| 27 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.449 |
0.054 |
| 28 |
257480_at |
AT1G15640
|
unknown |
0.448 |
-0.215 |
| 29 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.627 |
0.066 |
| 30 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
-0.548 |
0.338 |
| 31 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.526 |
-0.350 |
| 32 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.394 |
-0.153 |
| 33 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
-0.392 |
-0.053 |
| 34 |
245151_at |
AT2G47550
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.386 |
-0.197 |
| 35 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.361 |
-0.270 |
| 36 |
258075_at |
AT3G25900
|
ATHMT-1, HMT-1 |
-0.344 |
0.130 |
| 37 |
265208_at |
AT2G36690
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.332 |
0.099 |
| 38 |
251605_at |
AT3G57830
|
[Leucine-rich repeat protein kinase family protein] |
-0.311 |
0.064 |
| 39 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.304 |
-0.320 |
| 40 |
264294_at |
AT1G78750
|
[F-box/RNI-like superfamily protein] |
-0.298 |
0.042 |
| 41 |
261374_at |
AT1G52990
|
[thioredoxin family protein] |
-0.293 |
0.037 |
| 42 |
247175_at |
AT5G65280
|
GCL1, GCR2-like 1 |
-0.275 |
0.010 |
| 43 |
251466_at |
AT3G59340
|
unknown |
-0.265 |
-0.091 |
| 44 |
264794_at |
AT1G08670
|
[ENTH/VHS family protein] |
-0.263 |
-0.136 |
| 45 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.260 |
0.519 |
| 46 |
252836_at |
AT3G42070
|
unknown |
-0.255 |
0.009 |
| 47 |
249971_at |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
-0.250 |
-0.003 |
| 48 |
263963_at |
AT2G36080
|
ABS2, ABNORMAL SHOOT 2, NGAL1, NGATHA-Like 1 |
-0.233 |
-0.011 |
| 49 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.232 |
-0.174 |
| 50 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.228 |
-0.079 |
| 51 |
248577_at |
AT5G49870
|
[Mannose-binding lectin superfamily protein] |
-0.217 |
0.038 |
| 52 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.216 |
-0.062 |
| 53 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.216 |
-0.031 |
| 54 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
-0.214 |
0.280 |
| 55 |
256408_at |
AT1G66610
|
[TRAF-like superfamily protein] |
-0.205 |
-0.004 |