|
probeID |
AGICode |
Annotation |
Log2 signal ratio
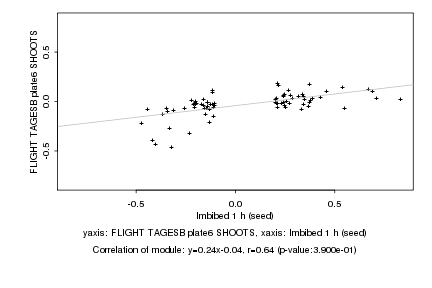
Imbibed 1 h (seed) |
Log2 signal ratio
FLIGHT TAGESB plate6 SHOOTS |
| 1 |
253113_at |
AT4G35985
|
[Senescence/dehydration-associated protein-related] |
0.829 |
0.030 |
| 2 |
257076_at |
AT3G19680
|
unknown |
0.710 |
0.034 |
| 3 |
252212_at |
AT3G50310
|
MAPKKK20, mitogen-activated protein kinase kinase kinase 20, MKKK20, MAPKK kinase 20 |
0.688 |
0.108 |
| 4 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.668 |
0.128 |
| 5 |
266269_at |
AT2G29480
|
ATGSTU2, glutathione S-transferase tau 2, GST20, GLUTATHIONE S-TRANSFERASE 20, GSTU2, glutathione S-transferase tau 2 |
0.546 |
-0.067 |
| 6 |
257654_at |
AT3G13310
|
DJC66, DNA J protein C66 |
0.537 |
0.147 |
| 7 |
250098_at |
AT5G17350
|
unknown |
0.456 |
0.109 |
| 8 |
255807_at |
AT4G10270
|
[Wound-responsive family protein] |
0.424 |
0.043 |
| 9 |
245757_at |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
0.386 |
0.037 |
| 10 |
259106_at |
AT3G05490
|
RALFL22, ralf-like 22 |
0.373 |
0.015 |
| 11 |
267238_at |
AT2G44130
|
KMD3, KISS ME DEADLY 3 |
0.368 |
0.173 |
| 12 |
249583_at |
AT5G37770
|
CML24, CALMODULIN-LIKE 24, TCH2, TOUCH 2 |
0.368 |
-0.006 |
| 13 |
250023_at |
AT5G18220
|
[O-Glycosyl hydrolases family 17 protein] |
0.363 |
-0.047 |
| 14 |
259803_at |
AT1G72150
|
PATL1, PATELLIN 1 |
0.347 |
0.026 |
| 15 |
245096_at |
AT2G40880
|
ATCYSA, cystatin A, CYSA, cystatin A, FL3-27 |
0.342 |
-0.023 |
| 16 |
264704_at |
AT1G70090
|
GATL9, GALACTURONOSYLTRANSFERASE-LIKE 9, LGT8, glucosyl transferase family 8 |
0.340 |
0.053 |
| 17 |
248981_at |
AT5G45110
|
ATNPR3, NPR3, NPR1-like protein 3 |
0.335 |
0.080 |
| 18 |
260803_at |
AT1G78340
|
ATGSTU22, glutathione S-transferase TAU 22, GSTU22, glutathione S-transferase TAU 22 |
0.331 |
-0.078 |
| 19 |
252671_at |
AT3G44190
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.315 |
0.054 |
| 20 |
251640_at |
AT3G57450
|
unknown |
0.284 |
0.033 |
| 21 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
0.275 |
0.061 |
| 22 |
258979_at |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
0.272 |
-0.014 |
| 23 |
254605_at |
AT4G18950
|
[Integrin-linked protein kinase family] |
0.266 |
0.116 |
| 24 |
256430_at |
AT3G11020
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2B, DRE/CRT-binding protein 2B |
0.256 |
0.005 |
| 25 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.247 |
-0.056 |
| 26 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.245 |
-0.040 |
| 27 |
264661_at |
AT1G09950
|
RAS1, RESPONSE TO ABA AND SALT 1 |
0.242 |
0.080 |
| 28 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
0.239 |
0.060 |
| 29 |
246018_at |
AT5G10695
|
unknown |
0.239 |
0.063 |
| 30 |
262155_at |
AT1G52420
|
[UDP-Glycosyltransferase superfamily protein] |
0.237 |
-0.007 |
| 31 |
245165_at |
AT2G33180
|
unknown |
0.227 |
-0.018 |
| 32 |
249253_at |
AT5G42060
|
[DEK, chromatin associated protein] |
0.216 |
0.170 |
| 33 |
260501_at |
AT2G41770
|
unknown |
0.210 |
-0.052 |
| 34 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.208 |
0.192 |
| 35 |
264089_at |
AT2G31200
|
ADF6, actin depolymerizing factor 6, ATADF6 |
0.208 |
-0.010 |
| 36 |
246193_at |
AT5G20880
|
[retrotransposon family] |
0.204 |
0.040 |
| 37 |
261031_at |
AT1G17360
|
unknown |
0.203 |
-0.018 |
| 38 |
260439_at |
AT1G68340
|
unknown |
0.199 |
0.000 |
| 39 |
256755_at |
AT3G25600
|
[Calcium-binding EF-hand family protein] |
0.198 |
0.023 |
| 40 |
244995_at |
ATCG00150
|
ATPI |
-0.476 |
-0.214 |
| 41 |
244973_at |
ATCG00690
|
PSBT, photosystem II reaction center protein T, PSBTC |
-0.443 |
-0.077 |
| 42 |
244999_at |
ATCG00190
|
RPOB, RNA polymerase subunit beta |
-0.421 |
-0.387 |
| 43 |
245005_at |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.405 |
-0.434 |
| 44 |
251258_at |
AT3G62170
|
VGDH2, VANGUARD 1 homolog 2 |
-0.372 |
-0.124 |
| 45 |
244933_at |
ATCG01070
|
NDHE |
-0.350 |
-0.065 |
| 46 |
245026_at |
ATCG00140
|
ATPH |
-0.343 |
-0.099 |
| 47 |
244934_at |
ATCG01080
|
NDHG |
-0.337 |
-0.269 |
| 48 |
256538_x_at |
AT3G32900
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT3G47260.1)] |
-0.325 |
-0.463 |
| 49 |
245008_at |
ATCG00360
|
YCF3 |
-0.316 |
-0.084 |
| 50 |
244985_at |
ATCG00810
|
RPL22, ribosomal protein L22 |
-0.259 |
-0.063 |
| 51 |
261235_x_at |
AT1G32840
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G14130.1)] |
-0.234 |
-0.316 |
| 52 |
261623_at |
AT1G01980
|
[ATSEC1A] |
-0.223 |
0.015 |
| 53 |
244982_at |
ATCG00780
|
RPL14, ribosomal protein L14 |
-0.212 |
-0.027 |
| 54 |
244964_at |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.210 |
-0.054 |
| 55 |
244986_at |
ATCG00820
|
RPS19, ribosomal protein S19 |
-0.207 |
-0.019 |
| 56 |
266865_at |
AT2G29980
|
AtFAD3, FAD3, fatty acid desaturase 3 |
-0.203 |
0.009 |
| 57 |
251252_at |
AT3G62230
|
DAF1, DUO1-activated F-box 1 |
-0.202 |
-0.022 |
| 58 |
266636_at |
AT2G35370
|
GDCH, glycine decarboxylase complex H |
-0.198 |
-0.019 |
| 59 |
244984_at |
ATCG00800
|
[RESISTANCE TO PSEUDOMONAS SYRINGAE 3 (RPS3)] |
-0.173 |
-0.027 |
| 60 |
252820_at |
AT3G42640
|
AHA8, H(+)-ATPase 8, HA8, H(+)-ATPase 8 |
-0.164 |
0.025 |
| 61 |
244963_at |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.161 |
-0.035 |
| 62 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.156 |
-0.065 |
| 63 |
244970_at |
ATCG00660
|
RPL20, ribosomal protein L20 |
-0.152 |
-0.126 |
| 64 |
248406_at |
AT5G51490
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.149 |
-0.070 |
| 65 |
244968_at |
ATCG00640
|
RPL33, ribosomal protein L33 |
-0.144 |
-0.008 |
| 66 |
254123_at |
AT4G24640
|
APPB1 |
-0.142 |
-0.001 |
| 67 |
258190_at |
AT3G29080
|
unknown |
-0.141 |
-0.048 |
| 68 |
255376_x_at |
AT4G03790
|
[gypsy-like retrotransposon family (Athila), has a 2.2e-286 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
-0.135 |
-0.205 |
| 69 |
256871_at |
AT3G26480
|
[Transducin family protein / WD-40 repeat family protein] |
-0.133 |
-0.073 |
| 70 |
267377_at |
AT2G26250
|
FDH, FIDDLEHEAD, KCS10, 3-ketoacyl-CoA synthase 10 |
-0.129 |
-0.025 |
| 71 |
263165_at |
AT1G03060
|
SPI, SPIRRIG |
-0.120 |
-0.024 |
| 72 |
258690_at |
AT3G07960
|
PIP5K6, phosphatidylinositol-4-phosphate 5-kinase 6 |
-0.119 |
0.092 |
| 73 |
252367_at |
AT3G48360
|
ATBT2, BT2, BTB and TAZ domain protein 2 |
-0.116 |
0.115 |
| 74 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.115 |
-0.149 |
| 75 |
258181_at |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-0.114 |
-0.059 |
| 76 |
256668_at |
AT3G32190
|
unknown |
-0.110 |
-0.033 |
| 77 |
251674_at |
AT3G57250
|
[Emsy N Terminus (ENT) domain-containing protein] |
-0.108 |
-0.018 |