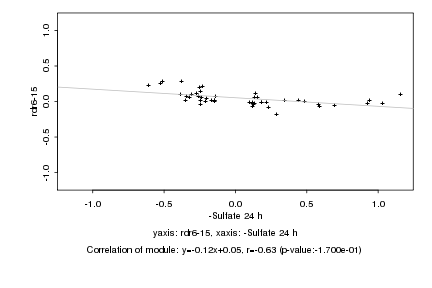
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
-Sulfate 24 h |
Log2 signal ratio
rdr6-15 |
| 1 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
1.154 |
0.103 |
| 2 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.026 |
-0.015 |
| 3 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.935 |
0.024 |
| 4 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.922 |
-0.019 |
| 5 |
256450_at |
AT1G75290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.691 |
-0.046 |
| 6 |
250475_at |
AT5G10180
|
AST68, ARABIDOPSIS SULFATE TRANSPORTER 68, SULTR2;1, sulfate transporter 2;1 |
0.588 |
-0.061 |
| 7 |
255105_at |
AT4G08620
|
SULTR1;1, sulphate transporter 1;1 |
0.577 |
-0.034 |
| 8 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.483 |
0.005 |
| 9 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.435 |
0.024 |
| 10 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.343 |
0.025 |
| 11 |
263231_at |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
0.285 |
-0.174 |
| 12 |
254343_at |
AT4G21990
|
APR3, APS reductase 3, ATAPR3, PRH-26, PRH26, PAPS REDUCTASE HOMOLOG 26 |
0.228 |
-0.081 |
| 13 |
251722_at |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
0.213 |
-0.011 |
| 14 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.177 |
-0.002 |
| 15 |
261475_at |
AT1G14550
|
[Peroxidase superfamily protein] |
0.176 |
-0.012 |
| 16 |
245254_at |
AT4G14680
|
APS3 |
0.149 |
0.069 |
| 17 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
0.139 |
0.124 |
| 18 |
249461_at |
AT5G39640
|
[Putative endonuclease or glycosyl hydrolase] |
0.132 |
-0.016 |
| 19 |
255999_at |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
0.128 |
0.058 |
| 20 |
256453_at |
AT1G75270
|
DHAR2, dehydroascorbate reductase 2 |
0.121 |
-0.041 |
| 21 |
247529_at |
AT5G61520
|
[Major facilitator superfamily protein] |
0.118 |
-0.014 |
| 22 |
249721_at |
AT5G24655
|
LSU4, RESPONSE TO LOW SULFUR 4 |
0.115 |
-0.068 |
| 23 |
266499_at |
AT2G06980
|
[pseudogene] |
0.114 |
-0.003 |
| 24 |
263853_at |
AT2G04440
|
[MutT/nudix family protein] |
0.096 |
-0.003 |
| 25 |
249866_at |
AT5G23010
|
IMS3, 2-ISOPROPYLMALATE SYNTHASE 3, MAM1, methylthioalkylmalate synthase 1 |
-0.611 |
0.228 |
| 26 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.530 |
0.256 |
| 27 |
257021_at |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
-0.512 |
0.283 |
| 28 |
255298_at |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
-0.388 |
0.101 |
| 29 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.383 |
0.288 |
| 30 |
254027_at |
AT4G25835
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.352 |
0.020 |
| 31 |
260745_at |
AT1G78370
|
ATGSTU20, glutathione S-transferase TAU 20, GSTU20, glutathione S-transferase TAU 20 |
-0.344 |
0.083 |
| 32 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
-0.326 |
0.058 |
| 33 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.309 |
0.103 |
| 34 |
254687_at |
AT4G13770
|
CYP83A1, cytochrome P450, family 83, subfamily A, polypeptide 1, REF2, REDUCED EPIDERMAL FLUORESCENCE 2 |
-0.274 |
0.125 |
| 35 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.261 |
0.076 |
| 36 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.256 |
0.200 |
| 37 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.246 |
0.147 |
| 38 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
-0.246 |
0.016 |
| 39 |
265615_at |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.246 |
-0.037 |
| 40 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.240 |
0.062 |
| 41 |
254862_at |
AT4G12030
|
BASS5, BILE ACID:SODIUM SYMPORTER FAMILY PROTEIN 5, BAT5, bile acid transporter 5 |
-0.232 |
0.216 |
| 42 |
254912_at |
AT4G11230
|
[Riboflavin synthase-like superfamily protein] |
-0.212 |
0.001 |
| 43 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.205 |
0.055 |
| 44 |
254013_at |
AT4G26050
|
PIRL8, plant intracellular ras group-related LRR 8 |
-0.172 |
0.019 |
| 45 |
257197_at |
AT3G23800
|
SBP3, selenium-binding protein 3 |
-0.149 |
0.020 |
| 46 |
266307_at |
AT2G27000
|
CYP705A8, cytochrome P450, family 705, subfamily A, polypeptide 8 |
-0.149 |
0.014 |
| 47 |
250738_at |
AT5G05730
|
AMT1, A-METHYL TRYPTOPHAN RESISTANT 1, ASA1, anthranilate synthase alpha subunit 1, JDL1, JASMONATE-INDUCED DEFECTIVE LATERAL ROOT 1, TRP5, TRYPTOPHAN BIOSYNTHESIS 5, WEI2, WEAK ETHYLENE INSENSITIVE 2 |
-0.149 |
0.016 |
| 48 |
251106_at |
AT5G01500
|
TAAC, thylakoid ATP/ADP carrier |
-0.145 |
0.083 |