|
probeID |
AGICode |
Annotation |
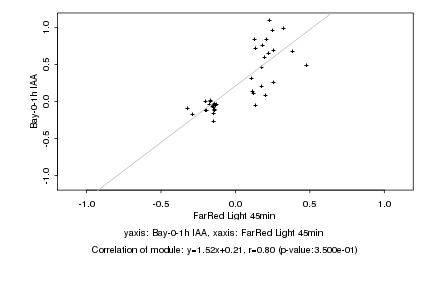
Log2 signal ratio
FarRed Light 45min |
Log2 signal ratio
Bay-0-1h IAA |
| 1 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
0.474 |
0.486 |
| 2 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
0.379 |
0.683 |
| 3 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
0.317 |
0.992 |
| 4 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
0.254 |
0.697 |
| 5 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
0.252 |
0.259 |
| 6 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
0.243 |
0.959 |
| 7 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
0.227 |
1.106 |
| 8 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
0.221 |
0.661 |
| 9 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
0.204 |
0.839 |
| 10 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
0.201 |
0.084 |
| 11 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
0.192 |
0.605 |
| 12 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
0.178 |
0.767 |
| 13 |
247778_at |
AT5G58750
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.170 |
0.211 |
| 14 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
0.169 |
0.459 |
| 15 |
248213_at |
AT5G53660
|
AtGRF7, growth-regulating factor 7, GRF7, growth-regulating factor 7 |
0.130 |
-0.042 |
| 16 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
0.130 |
0.720 |
| 17 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
0.128 |
0.841 |
| 18 |
265448_at |
AT2G46590
|
Dof-type zinc finger DNA-binding family protein |
0.116 |
0.121 |
| 19 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
0.112 |
0.140 |
| 20 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
0.107 |
0.316 |
| 21 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
-0.324 |
-0.088 |
| 22 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
-0.293 |
-0.175 |
| 23 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
-0.205 |
0.005 |
| 24 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
-0.204 |
-0.118 |
| 25 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
-0.198 |
-0.117 |
| 26 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.175 |
-0.033 |
| 27 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
-0.174 |
0.001 |
| 28 |
267336_at |
AT2G19310
|
[HSP20-like chaperones superfamily protein] |
-0.170 |
0.027 |
| 29 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
-0.158 |
-0.056 |
| 30 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
-0.153 |
-0.158 |
| 31 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
-0.151 |
-0.266 |
| 32 |
252519_at |
AT3G46360
|
unknown |
-0.150 |
-0.044 |
| 33 |
262811_at |
AT1G11700
|
unknown |
-0.148 |
-0.073 |
| 34 |
256518_at |
AT1G66080
|
unknown |
-0.146 |
-0.108 |
| 35 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
-0.146 |
-0.102 |
| 36 |
258830_at |
AT3G07090
|
[PPPDE putative thiol peptidase family protein] |
-0.146 |
-0.030 |
| 37 |
254211_at |
AT4G23570
|
SGT1A |
-0.143 |
-0.019 |
| 38 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
-0.136 |
-0.041 |
| 39 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
-0.128 |
-0.034 |