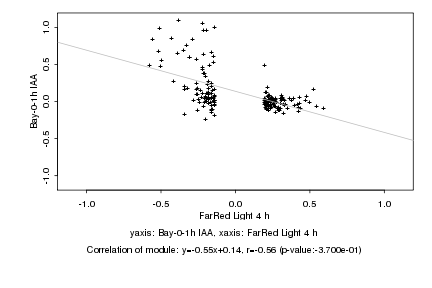
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
FarRed Light 4 h |
Log2 signal ratio
Bay-0-1h IAA |
| 1 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.585 |
-0.087 |
| 2 |
250533_at |
AT5G08640
|
ATFLS1, FLS, FLAVONOL SYNTHASE, FLS1, flavonol synthase 1 |
0.538 |
-0.054 |
| 3 |
252661_at |
AT3G44450
|
unknown |
0.522 |
0.169 |
| 4 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.496 |
-0.012 |
| 5 |
252010_at |
AT3G52740
|
unknown |
0.474 |
0.079 |
| 6 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.467 |
0.019 |
| 7 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.433 |
-0.082 |
| 8 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.426 |
-0.018 |
| 9 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
0.426 |
0.058 |
| 10 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.420 |
-0.071 |
| 11 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.420 |
-0.133 |
| 12 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.415 |
-0.028 |
| 13 |
265892_at |
AT2G15020
|
unknown |
0.396 |
-0.047 |
| 14 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.389 |
0.052 |
| 15 |
248625_at |
AT5G48880
|
KAT5, 3-KETO-ACYL-COENZYME A THIOLASE 5, PKT1, PEROXISOMAL-3-KETO-ACYL-COA THIOLASE 1, PKT2, peroxisomal 3-keto-acyl-CoA thiolase 2 |
0.372 |
0.015 |
| 16 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.360 |
0.042 |
| 17 |
250207_at |
AT5G13930
|
ATCHS, CHS, CHALCONE SYNTHASE, TT4, TRANSPARENT TESTA 4 |
0.343 |
-0.090 |
| 18 |
247779_at |
AT5G58760
|
DDB2, damaged DNA binding 2 |
0.334 |
-0.030 |
| 19 |
245306_at |
AT4G14690
|
ELIP2, EARLY LIGHT-INDUCIBLE PROTEIN 2 |
0.329 |
-0.046 |
| 20 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.328 |
0.017 |
| 21 |
261907_at |
AT1G65060
|
4CL3, 4-coumarate:CoA ligase 3 |
0.321 |
-0.039 |
| 22 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.321 |
-0.154 |
| 23 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
0.319 |
0.037 |
| 24 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
0.313 |
0.063 |
| 25 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
0.307 |
0.026 |
| 26 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
0.306 |
0.093 |
| 27 |
249622_at |
AT5G37550
|
unknown |
0.302 |
0.048 |
| 28 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.296 |
-0.085 |
| 29 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.296 |
-0.023 |
| 30 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.291 |
-0.116 |
| 31 |
247463_at |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
0.287 |
-0.076 |
| 32 |
255877_at |
AT2G40460
|
[Major facilitator superfamily protein] |
0.283 |
-0.120 |
| 33 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.282 |
-0.092 |
| 34 |
262626_at |
AT1G06430
|
FTSH8, FTSH protease 8 |
0.273 |
-0.012 |
| 35 |
266257_at |
AT2G27820
|
ADT3, arogenate dehydratase 3, PD1, prephenate dehydratase 1 |
0.268 |
0.001 |
| 36 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.267 |
-0.061 |
| 37 |
263545_at |
AT2G21560
|
unknown |
0.264 |
-0.148 |
| 38 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.263 |
0.039 |
| 39 |
258696_at |
AT3G09650
|
CRM3, HCF152, HIGH CHLOROPHYLL FLUORESCENCE 152 |
0.263 |
0.042 |
| 40 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.260 |
0.013 |
| 41 |
252073_at |
AT3G51750
|
unknown |
0.259 |
0.004 |
| 42 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.257 |
0.014 |
| 43 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.257 |
-0.055 |
| 44 |
248347_at |
AT5G52250
|
EFO1, EARLY FLOWERING BY OVEREXPRESSION 1, RUP1, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 |
0.255 |
-0.029 |
| 45 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.252 |
0.029 |
| 46 |
263122_at |
AT1G78510
|
AtSPS1, SPS1, solanesyl diphosphate synthase 1 |
0.251 |
-0.018 |
| 47 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
0.251 |
0.002 |
| 48 |
261958_at |
AT1G64500
|
[Glutaredoxin family protein] |
0.246 |
0.023 |
| 49 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.246 |
0.046 |
| 50 |
246411_at |
AT1G57770
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
0.244 |
0.042 |
| 51 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.240 |
-0.074 |
| 52 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.238 |
0.041 |
| 53 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.236 |
0.056 |
| 54 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
0.233 |
-0.001 |
| 55 |
248537_at |
AT5G50100
|
[Putative thiol-disulphide oxidoreductase DCC] |
0.230 |
-0.083 |
| 56 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
0.230 |
-0.031 |
| 57 |
251605_at |
AT3G57830
|
[Leucine-rich repeat protein kinase family protein] |
0.230 |
0.012 |
| 58 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
0.230 |
-0.044 |
| 59 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.229 |
-0.030 |
| 60 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.227 |
0.084 |
| 61 |
255793_at |
AT2G33250
|
unknown |
0.226 |
0.040 |
| 62 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
0.222 |
-0.059 |
| 63 |
262634_at |
AT1G06690
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.220 |
-0.033 |
| 64 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.219 |
-0.065 |
| 65 |
251725_at |
AT3G56260
|
unknown |
0.219 |
-0.113 |
| 66 |
250256_at |
AT5G13650
|
SVR3, SUPPRESSOR OF VARIEGATION 3 |
0.218 |
0.030 |
| 67 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
0.217 |
0.085 |
| 68 |
259970_at |
AT1G76570
|
AtLHCB7, LHCB7, light-harvesting complex B7 |
0.217 |
0.071 |
| 69 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.216 |
-0.013 |
| 70 |
263287_at |
AT2G36145
|
unknown |
0.211 |
-0.007 |
| 71 |
255594_at |
AT4G01660
|
ABC1, ABC transporter 1, ATABC1, ABC transporter 1, ATATH10 |
0.211 |
-0.010 |
| 72 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
0.210 |
0.197 |
| 73 |
251309_at |
AT3G61220
|
SDR1, short-chain dehydrogenase/reductase 1 |
0.209 |
0.071 |
| 74 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
0.208 |
0.133 |
| 75 |
257949_at |
AT3G21750
|
UGT71B1, UDP-glucosyl transferase 71B1 |
0.206 |
-0.090 |
| 76 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.206 |
-0.054 |
| 77 |
249769_at |
AT5G24120
|
ATSIG5, SIGMA FACTOR 5, SIG5, SIGMA FACTOR 5, SIGE, sigma factor E |
0.206 |
-0.051 |
| 78 |
260955_at |
AT1G06000
|
[UDP-Glycosyltransferase superfamily protein] |
0.205 |
-0.084 |
| 79 |
249244_at |
AT5G42270
|
FTSH5, VAR1, VARIEGATED 1 |
0.204 |
0.007 |
| 80 |
260300_at |
AT1G80340
|
ATGA3OX2, ARABIDOPSIS THALIANA GIBBERELLIN-3-OXIDASE 2, GA3OX2, gibberellin 3-oxidase 2, GA4H |
0.202 |
-0.100 |
| 81 |
263386_at |
AT2G40150
|
TBL28, TRICHOME BIREFRINGENCE-LIKE 28 |
0.202 |
-0.026 |
| 82 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.201 |
-0.024 |
| 83 |
253688_at |
AT4G29590
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.201 |
0.040 |
| 84 |
256097_at |
AT1G13670
|
unknown |
0.198 |
0.132 |
| 85 |
248204_at |
AT5G54280
|
ATM2, myosin 2, ATM4, ARABIDOPSIS THALIANA MYOSIN 4, ATMYOS1, ARABIDOPSIS THALIANA MYOSIN 1 |
0.198 |
-0.077 |
| 86 |
253835_at |
AT4G27820
|
BGLU9, beta glucosidase 9 |
0.198 |
-0.021 |
| 87 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.196 |
0.006 |
| 88 |
249677_at |
AT5G35970
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.195 |
0.054 |
| 89 |
252411_at |
AT3G47430
|
PEX11B, peroxin 11B |
0.193 |
-0.004 |
| 90 |
257262_at |
AT3G21890
|
BBX31, B-box domain protein 31 |
0.192 |
0.021 |
| 91 |
262945_at |
AT1G79510
|
[Uncharacterized conserved protein (DUF2358)] |
0.190 |
-0.028 |
| 92 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
0.190 |
0.495 |
| 93 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.579 |
0.486 |
| 94 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.558 |
0.841 |
| 95 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.518 |
0.683 |
| 96 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.513 |
0.992 |
| 97 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.507 |
0.473 |
| 98 |
252765_at |
AT3G42800
|
unknown |
-0.498 |
0.554 |
| 99 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.435 |
0.860 |
| 100 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.418 |
0.271 |
| 101 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.394 |
0.649 |
| 102 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.388 |
1.106 |
| 103 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.350 |
0.697 |
| 104 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.348 |
0.208 |
| 105 |
264021_at |
AT2G21200
|
SAUR7, SMALL AUXIN UPREGULATED RNA 7 |
-0.345 |
0.173 |
| 106 |
255608_at |
AT4G01140
|
unknown |
-0.343 |
-0.168 |
| 107 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.332 |
0.767 |
| 108 |
246562_at |
AT5G15580
|
LNG1, LONGIFOLIA1, TRM2, TON1 Recruiting Motif 2 |
-0.324 |
0.179 |
| 109 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.311 |
0.599 |
| 110 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.309 |
0.605 |
| 111 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.290 |
0.839 |
| 112 |
266688_at |
AT2G19660
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.286 |
0.018 |
| 113 |
258552_at |
AT3G07010
|
[Pectin lyase-like superfamily protein] |
-0.268 |
0.098 |
| 114 |
262236_at |
AT1G48330
|
unknown |
-0.266 |
0.174 |
| 115 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.263 |
0.251 |
| 116 |
248801_at |
AT5G47370
|
HAT2 |
-0.263 |
0.567 |
| 117 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.258 |
-0.117 |
| 118 |
256621_at |
AT3G24450
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.258 |
0.098 |
| 119 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.257 |
0.179 |
| 120 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.254 |
0.030 |
| 121 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.244 |
-0.012 |
| 122 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
-0.236 |
0.155 |
| 123 |
260297_at |
AT1G80280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.229 |
0.062 |
| 124 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.227 |
1.053 |
| 125 |
263690_at |
AT1G26960
|
AtHB23, homeobox protein 23, HB23, homeobox protein 23 |
-0.226 |
0.117 |
| 126 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.225 |
0.470 |
| 127 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.222 |
0.438 |
| 128 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.221 |
0.386 |
| 129 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.219 |
0.968 |
| 130 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.219 |
-0.055 |
| 131 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.219 |
0.640 |
| 132 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.211 |
0.047 |
| 133 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.210 |
0.383 |
| 134 |
259351_at |
AT3G05150
|
[Major facilitator superfamily protein] |
-0.209 |
-0.004 |
| 135 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.207 |
0.343 |
| 136 |
249807_at |
AT5G23870
|
[Pectinacetylesterase family protein] |
-0.207 |
0.074 |
| 137 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.206 |
-0.232 |
| 138 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
-0.205 |
0.001 |
| 139 |
254954_at |
AT4G10910
|
unknown |
-0.201 |
0.012 |
| 140 |
246997_at |
AT5G67390
|
unknown |
-0.199 |
0.046 |
| 141 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.198 |
0.959 |
| 142 |
263151_at |
AT1G54120
|
unknown |
-0.196 |
0.109 |
| 143 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.192 |
0.238 |
| 144 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.190 |
0.116 |
| 145 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.186 |
0.181 |
| 146 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.184 |
0.105 |
| 147 |
258367_at |
AT3G14370
|
WAG2 |
-0.184 |
0.275 |
| 148 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.183 |
0.049 |
| 149 |
250810_at |
AT5G05090
|
[Homeodomain-like superfamily protein] |
-0.183 |
0.124 |
| 150 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.182 |
0.066 |
| 151 |
249278_at |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-0.181 |
0.029 |
| 152 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.181 |
-0.023 |
| 153 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.177 |
0.490 |
| 154 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.172 |
-0.007 |
| 155 |
257547_at |
AT3G13000
|
unknown |
-0.167 |
0.121 |
| 156 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.167 |
-0.113 |
| 157 |
254373_at |
AT4G21730
|
[expressed protein] |
-0.167 |
0.058 |
| 158 |
250108_at |
AT5G15150
|
ATHB-3, homeobox 3, ATHB3, HAT7, HOMEOBOX FROM ARABIDOPSIS THALIANA 7, HB-3, homeobox 3 |
-0.166 |
-0.043 |
| 159 |
247903_at |
AT5G57340
|
unknown |
-0.164 |
0.213 |
| 160 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.164 |
0.044 |
| 161 |
257113_at |
AT3G20130
|
CYP705A22, cytochrome P450, family 705, subfamily A, polypeptide 22, GPS1, gravity persistence signal 1 |
-0.164 |
-0.141 |
| 162 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.162 |
-0.107 |
| 163 |
261825_at |
AT1G11545
|
XTH8, xyloglucan endotransglucosylase/hydrolase 8 |
-0.162 |
0.048 |
| 164 |
248623_at |
AT5G49170
|
unknown |
-0.161 |
0.254 |
| 165 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.161 |
0.664 |
| 166 |
258528_at |
AT3G06770
|
[Pectin lyase-like superfamily protein] |
-0.158 |
0.154 |
| 167 |
260806_at |
AT1G78260
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.155 |
-0.096 |
| 168 |
264023_at |
AT2G21195
|
unknown |
-0.154 |
0.002 |
| 169 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.154 |
0.535 |
| 170 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.154 |
0.611 |
| 171 |
251774_at |
AT3G55840
|
[Hs1pro-1 protein] |
-0.150 |
0.063 |
| 172 |
248912_at |
AT5G45670
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.148 |
0.049 |
| 173 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
-0.146 |
-0.013 |
| 174 |
251864_at |
AT3G54920
|
PMR6, powdery mildew resistant 6 |
-0.145 |
0.075 |
| 175 |
257071_at |
AT3G28180
|
ATCSLC04, Cellulose-synthase-like C4, ATCSLC4, CSLC04, CSLC04, Cellulose-synthase-like C4, CSLC4, CELLULOSE-SYNTHASE LIKE C4 |
-0.144 |
0.093 |
| 176 |
260434_at |
AT1G68330
|
unknown |
-0.144 |
0.166 |
| 177 |
252217_at |
AT3G50140
|
unknown |
-0.143 |
0.027 |
| 178 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.143 |
1.006 |
| 179 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.143 |
0.023 |
| 180 |
267210_at |
AT2G30920
|
ATCOQ3, coenzyme Q 3, COQ3, COQ3, coenzyme Q 3, EMB3002, embryo defective 3002 |
-0.142 |
-0.052 |
| 181 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.142 |
0.021 |
| 182 |
257618_at |
AT3G24715
|
[Protein kinase superfamily protein with octicosapeptide/Phox/Bem1p domain] |
-0.142 |
-0.030 |
| 183 |
265688_at |
AT2G24300
|
[Calmodulin-binding protein] |
-0.141 |
-0.180 |