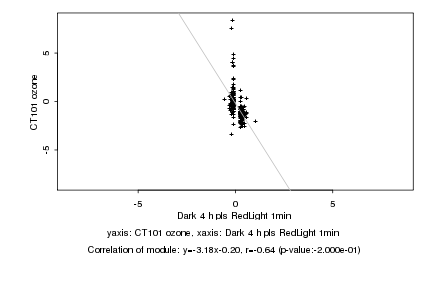
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Dark 4 h pls RedLight 1min |
Log2 signal ratio
CT101 ozone |
| 1 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.984 |
-2.017 |
| 2 |
260877_at |
AT1G21500
|
unknown |
0.560 |
-1.119 |
| 3 |
248684_at |
AT5G48485
|
DIR1, DEFECTIVE IN INDUCED RESISTANCE 1 |
0.544 |
0.333 |
| 4 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
0.525 |
-1.568 |
| 5 |
252441_at |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
0.524 |
-1.224 |
| 6 |
251814_at |
AT3G54890
|
LHCA1, photosystem I light harvesting complex gene 1 |
0.491 |
-1.320 |
| 7 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
0.431 |
-2.487 |
| 8 |
249876_at |
AT5G23060
|
CaS, calcium sensing receptor |
0.429 |
-2.225 |
| 9 |
255248_at |
AT4G05180
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-2, photosystem II subunit Q-2, PSII-Q |
0.418 |
-1.058 |
| 10 |
260542_at |
AT2G43560
|
[FKBP-like peptidyl-prolyl cis-trans isomerase family protein] |
0.417 |
-0.433 |
| 11 |
249645_at |
AT5G36910
|
THI2.2, thionin 2.2 |
0.413 |
-1.549 |
| 12 |
259840_at |
AT1G52230
|
PSAH-2, PHOTOSYSTEM I SUBUNIT H-2, PSAH2, photosystem I subunit H2, PSI-H |
0.411 |
-1.350 |
| 13 |
256309_at |
AT1G30380
|
PSAK, photosystem I subunit K |
0.409 |
-0.930 |
| 14 |
259491_at |
AT1G15820
|
CP24, LHCB6, light harvesting complex photosystem II subunit 6 |
0.409 |
-1.476 |
| 15 |
248151_at |
AT5G54270
|
LHCB3, light-harvesting chlorophyll B-binding protein 3, LHCB3*1 |
0.403 |
-1.377 |
| 16 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
0.385 |
-0.816 |
| 17 |
250006_at |
AT5G18660
|
PCB2, PALE-GREEN AND CHLOROPHYLL B REDUCED 2 |
0.385 |
-0.815 |
| 18 |
255457_at |
AT4G02770
|
PSAD-1, photosystem I subunit D-1 |
0.378 |
-1.424 |
| 19 |
256979_at |
AT3G21055
|
PSBTN, photosystem II subunit T |
0.371 |
-1.033 |
| 20 |
253009_at |
AT4G37930
|
SHM1, serine transhydroxymethyltransferase 1, SHMT1, SERINE HYDROXYMETHYLTRANSFERASE 1, STM, SERINE TRANSHYDROXYMETHYLTRANSFERASE |
0.367 |
-0.596 |
| 21 |
255046_at |
AT4G09650
|
ATPD, ATP synthase delta-subunit gene, PDE332, PIGMENT DEFECTIVE 332 |
0.367 |
-1.520 |
| 22 |
261746_at |
AT1G08380
|
PSAO, photosystem I subunit O |
0.366 |
-1.094 |
| 23 |
251519_at |
AT3G59400
|
GUN4, GENOMES UNCOUPLED 4 |
0.361 |
-2.557 |
| 24 |
263760_at |
AT2G21280
|
ATSULA, GC1, GIANT CHLOROPLAST 1, SULA |
0.360 |
-0.616 |
| 25 |
250243_at |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
0.349 |
-1.773 |
| 26 |
259460_at |
AT1G44000
|
unknown |
0.347 |
-1.744 |
| 27 |
258607_at |
AT3G02730
|
ATF1, TRXF1, thioredoxin F-type 1 |
0.336 |
-1.090 |
| 28 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.334 |
-1.164 |
| 29 |
263841_at |
AT2G36870
|
AtXTH32, XTH32, xyloglucan endotransglucosylase/hydrolase 32 |
0.333 |
-2.363 |
| 30 |
264839_at |
AT1G03630
|
POR C, protochlorophyllide oxidoreductase C, PORC |
0.330 |
-1.524 |
| 31 |
255886_at |
AT1G20340
|
DRT112, DNA-DAMAGE-REPAIR/TOLERATION PROTEIN 112, PETE2, PLASTOCYANIN 2 |
0.329 |
-1.029 |
| 32 |
253738_at |
AT4G28750
|
PSAE-1, PSA E1 KNOCKOUT |
0.327 |
-0.975 |
| 33 |
258333_at |
AT3G16000
|
MFP1, MAR binding filament-like protein 1 |
0.326 |
-1.407 |
| 34 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.321 |
-1.285 |
| 35 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.320 |
-2.134 |
| 36 |
256015_at |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
0.316 |
-1.943 |
| 37 |
252430_at |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
0.314 |
-1.233 |
| 38 |
257932_at |
AT3G17040
|
HCF107, high chlorophyll fluorescent 107 |
0.307 |
-1.569 |
| 39 |
262970_at |
AT1G75690
|
LQY1, LOW QUANTUM YIELD OF PHOTOSYSTEM II 1 |
0.305 |
-0.896 |
| 40 |
259892_at |
AT1G72610
|
ATGER1, A. THALIANA GERMIN-LIKE PROTEIN 1, GER1, germin-like protein 1, GLP1, GERMIN-LIKE PROTEIN 1 |
0.299 |
-2.517 |
| 41 |
254790_at |
AT4G12800
|
PSAL, photosystem I subunit l |
0.298 |
-0.884 |
| 42 |
249927_at |
AT5G19220
|
ADG2, ADP GLUCOSE PYROPHOSPHORYLASE 2, APL1, ADP glucose pyrophosphorylase large subunit 1 |
0.294 |
-0.474 |
| 43 |
245165_at |
AT2G33180
|
unknown |
0.293 |
-1.162 |
| 44 |
255675_at |
AT4G00480
|
ATMYC1, myc1 |
0.292 |
-0.991 |
| 45 |
254398_at |
AT4G21280
|
PSBQ, PHOTOSYSTEM II SUBUNIT Q, PSBQ-1, PHOTOSYSTEM II SUBUNIT Q-1, PSBQA, photosystem II subunit QA |
0.292 |
-1.006 |
| 46 |
262736_at |
AT1G28570
|
[SGNH hydrolase-type esterase superfamily protein] |
0.290 |
-2.191 |
| 47 |
253387_at |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
0.290 |
-1.080 |
| 48 |
263761_at |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
0.289 |
-2.041 |
| 49 |
263114_at |
AT1G03130
|
PSAD-2, photosystem I subunit D-2 |
0.288 |
-1.749 |
| 50 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
0.286 |
-2.300 |
| 51 |
261751_at |
AT1G76080
|
ATCDSP32, ARABIDOPSIS THALIANA CHLOROPLASTIC DROUGHT-INDUCED STRESS PROTEIN OF 32 KD, CDSP32, chloroplastic drought-induced stress protein of 32 kD |
0.283 |
-0.931 |
| 52 |
261422_at |
AT1G18730
|
NDF6, NDH dependent flow 6, PnsB4, Photosynthetic NDH subcomplex B 4 |
0.282 |
-0.687 |
| 53 |
254623_at |
AT4G18480
|
CH-42, CHLORINA 42, CH42, CHLORINA 42, CHL11, CHLI-1, CHLI1 |
0.280 |
-1.012 |
| 54 |
258956_at |
AT3G01440
|
PnsL3, Photosynthetic NDH subcomplex L 3, PQL1, PsbQ-like 1, PQL2, PsbQ-like 2 |
0.279 |
-1.697 |
| 55 |
251664_at |
AT3G56940
|
ACSF, CHL27, CRD1, COPPER RESPONSE DEFECT 1 |
0.277 |
-1.744 |
| 56 |
262283_at |
AT1G68590
|
PSRP3/1, plastid-specific ribosomal protein 3/1 |
0.276 |
-0.828 |
| 57 |
260547_at |
AT2G43550
|
[Scorpion toxin-like knottin superfamily protein] |
0.275 |
-1.220 |
| 58 |
262557_at |
AT1G31330
|
PSAF, photosystem I subunit F |
0.272 |
-1.040 |
| 59 |
258589_at |
AT3G04290
|
ATLTL1, LTL1, Li-tolerant lipase 1 |
0.270 |
-0.816 |
| 60 |
252130_at |
AT3G50820
|
OEC33, OXYGEN EVOLVING COMPLEX SUBUNIT 33 KDA, PSBO-2, PHOTOSYSTEM II SUBUNIT O-2, PSBO2, photosystem II subunit O-2 |
0.268 |
-1.452 |
| 61 |
262168_at |
AT1G74730
|
unknown |
0.265 |
-0.879 |
| 62 |
255447_at |
AT4G02790
|
EMB3129, EMBRYO DEFECTIVE 3129 |
0.264 |
-1.034 |
| 63 |
245195_at |
AT1G67740
|
PSBY, photosystem II BY, YCF32 |
0.262 |
-1.492 |
| 64 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.262 |
0.419 |
| 65 |
245123_at |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
0.260 |
-1.557 |
| 66 |
249524_at |
AT5G38520
|
[alpha/beta-Hydrolases superfamily protein] |
0.259 |
-1.063 |
| 67 |
262645_at |
AT1G62750
|
ATSCO1, SNOWY COTYLEDON 1, ATSCO1/CPEF-G, SCO1, SNOWY COTYLEDON 1 |
0.257 |
-1.462 |
| 68 |
245213_at |
AT1G44575
|
CP22, NPQ4, NONPHOTOCHEMICAL QUENCHING 4, PSBS, PHOTOSYSTEM II SUBUNIT S |
0.257 |
-0.764 |
| 69 |
267526_at |
AT2G30570
|
PSBW, photosystem II reaction center W |
0.257 |
-1.151 |
| 70 |
261769_at |
AT1G76100
|
PETE1, plastocyanin 1 |
0.255 |
-1.479 |
| 71 |
261197_at |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
0.254 |
-1.495 |
| 72 |
252463_at |
AT3G47070
|
[LOCATED IN: thylakoid, chloroplast thylakoid membrane, chloroplast, chloroplast envelope] |
0.253 |
-0.929 |
| 73 |
264360_at |
AT1G03310
|
AtBE2, ATISA2, ARABIDOPSIS THALIANA ISOAMYLASE 2, BE2, BRANCHING ENZYME 2, DBE1, debranching enzyme 1, ISA2 |
0.251 |
-2.123 |
| 74 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
0.250 |
-1.285 |
| 75 |
264185_at |
AT1G54780
|
AtTLP18.3, TLP18.3, thylakoid lumen protein 18.3 |
0.249 |
-1.468 |
| 76 |
248920_at |
AT5G45930
|
CHL I2, CHLI-2, CHLI2, magnesium chelatase i2 |
0.249 |
-0.983 |
| 77 |
258285_at |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
0.249 |
-0.600 |
| 78 |
249007_at |
AT5G44650
|
AtCEST, Arabidopsis thaliana chloroplast protein-enhancing stress tolerance, CEST, chloroplast protein-enhancing stress tolerance, Y3IP1, Ycf3-interacting protein 1 |
0.248 |
-0.763 |
| 79 |
246268_at |
AT1G31800
|
CYP97A3, cytochrome P450, family 97, subfamily A, polypeptide 3, LUT5, LUTEIN DEFICIENT 5 |
0.248 |
-0.803 |
| 80 |
253860_at |
AT4G27700
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.247 |
-1.999 |
| 81 |
248028_at |
AT5G55620
|
unknown |
0.244 |
-2.142 |
| 82 |
263142_at |
AT1G65230
|
[Uncharacterized conserved protein (DUF2358)] |
0.244 |
-1.555 |
| 83 |
257311_at |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
0.244 |
-1.404 |
| 84 |
259161_at |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
0.242 |
-0.987 |
| 85 |
266979_at |
AT2G39470
|
PnsL1, Photosynthetic NDH subcomplex L 1, PPL2, PsbP-like protein 2 |
0.241 |
-1.011 |
| 86 |
256469_at |
AT1G32540
|
LOL1, lsd one like 1 |
0.239 |
0.056 |
| 87 |
261788_at |
AT1G15980
|
NDF1, NDH-dependent cyclic electron flow 1, NDH48, NAD(P)H DEHYDROGENASE SUBUNIT 48, PnsB1, Photosynthetic NDH subcomplex B 1 |
0.237 |
-1.137 |
| 88 |
247320_at |
AT5G64040
|
PSAN |
0.237 |
-1.117 |
| 89 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.236 |
-0.580 |
| 90 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
0.235 |
-2.258 |
| 91 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.235 |
-2.143 |
| 92 |
265033_at |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
0.234 |
-1.309 |
| 93 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
0.233 |
-2.584 |
| 94 |
255078_at |
AT4G09010
|
APX4, ascorbate peroxidase 4, TL29, thylakoid lumen 29 |
0.232 |
-0.672 |
| 95 |
251031_at |
AT5G02120
|
OHP, one helix protein, PDE335, PIGMENT DEFECTIVE 335 |
0.229 |
-1.202 |
| 96 |
265374_at |
AT2G06520
|
PSBX, photosystem II subunit X |
0.229 |
-1.201 |
| 97 |
259838_at |
AT1G52220
|
CURT1C, CURVATURE THYLAKOID 1C |
0.226 |
-0.699 |
| 98 |
258895_at |
AT3G05600
|
[alpha/beta-Hydrolases superfamily protein] |
0.226 |
0.433 |
| 99 |
262879_at |
AT1G64860
|
RPOD1, SIG1, RNA POLYMERASE SIGMA SUBUNIT 1, SIG2, RNApolymerase sigma subunit 2, SIGA, sigma factor A, SIGB |
0.224 |
-2.009 |
| 100 |
254970_at |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
0.223 |
-1.245 |
| 101 |
251741_at |
AT3G56040
|
UGP3, UDP-glucose pyrophosphorylase 3 |
0.223 |
-1.857 |
| 102 |
247073_at |
AT5G66570
|
MSP-1, MANGANESE-STABILIZING PROTEIN 1, OE33, OXYGEN EVOLVING COMPLEX 33 KILODALTON PROTEIN, OEE1, 33 KDA OXYGEN EVOLVING POLYPEPTIDE 1, OEE33, OXYGEN EVOLVING ENHANCER PROTEIN 33, PSBO-1, PS II OXYGEN-EVOLVING COMPLEX 1, PSBO1, PS II oxygen-evolving complex 1 |
0.223 |
-1.098 |
| 103 |
255440_at |
AT4G02530
|
[chloroplast thylakoid lumen protein] |
0.220 |
-0.449 |
| 104 |
262632_at |
AT1G06680
|
OE23, OXYGEN EVOLVING COMPLEX SUBUNIT 23 KDA, OEE2, OXYGEN-EVOLVING ENHANCER PROTEIN 2, PSBP-1, photosystem II subunit P-1, PSII-P, PHOTOSYSTEM II SUBUNIT P |
0.220 |
-1.248 |
| 105 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.220 |
-1.382 |
| 106 |
250133_at |
AT5G16400
|
ATF2, TRXF2, thioredoxin F2 |
0.218 |
-1.032 |
| 107 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.217 |
-1.920 |
| 108 |
252929_at |
AT4G38970
|
AtFBA2, FBA2, fructose-bisphosphate aldolase 2 |
0.217 |
-1.099 |
| 109 |
258804_at |
AT3G04760
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.216 |
-1.210 |
| 110 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.216 |
1.162 |
| 111 |
266672_at |
AT2G29650
|
ANTR1, anion transporter 1, PHT4;1, phosphate transporter 4;1 |
0.216 |
-1.211 |
| 112 |
249120_at |
AT5G43750
|
NDH18, NAD(P)H dehydrogenase 18, PnsB5, Photosynthetic NDH subcomplex B 5 |
0.216 |
-0.791 |
| 113 |
254102_at |
AT4G25050
|
ACP4, acyl carrier protein 4 |
0.215 |
-0.653 |
| 114 |
252318_at |
AT3G48730
|
GSA2, glutamate-1-semialdehyde 2,1-aminomutase 2 |
0.214 |
-0.431 |
| 115 |
262376_at |
AT1G72970
|
EDA17, embryo sac development arrest 17, HTH, HOTHEAD |
0.212 |
-1.697 |
| 116 |
259799_at |
AT1G72290
|
[Kunitz family trypsin and protease inhibitor protein] |
-0.572 |
0.225 |
| 117 |
248435_at |
AT5G51210
|
OLEO3, oleosin3 |
-0.331 |
-0.710 |
| 118 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
-0.306 |
-0.308 |
| 119 |
245233_at |
AT4G25580
|
[CAP160 protein] |
-0.306 |
0.602 |
| 120 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.302 |
0.655 |
| 121 |
255905_at |
AT1G17810
|
BETA-TIP, beta-tonoplast intrinsic protein |
-0.299 |
-0.147 |
| 122 |
251012_at |
AT5G02580
|
unknown |
-0.279 |
0.225 |
| 123 |
262452_at |
AT1G11210
|
unknown |
-0.259 |
-0.914 |
| 124 |
262198_at |
AT1G53830
|
ATPME2, pectin methylesterase 2, PME2, pectin methylesterase 2 |
-0.257 |
-0.219 |
| 125 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.252 |
1.004 |
| 126 |
250214_at |
AT5G13870
|
EXGT-A4, endoxyloglucan transferase A4, XTH5, xyloglucan endotransglucosylase/hydrolase 5 |
-0.241 |
-1.269 |
| 127 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.239 |
-0.456 |
| 128 |
259935_at |
AT1G71250
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.225 |
0.530 |
| 129 |
256114_at |
AT1G16850
|
unknown |
-0.224 |
-3.307 |
| 130 |
263894_at |
AT2G21910
|
CYP96A5, cytochrome P450, family 96, subfamily A, polypeptide 5 |
-0.222 |
7.618 |
| 131 |
260213_at |
AT1G74490
|
[Protein kinase superfamily protein] |
-0.221 |
0.213 |
| 132 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.211 |
-0.131 |
| 133 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
-0.205 |
0.602 |
| 134 |
253522_at |
AT4G31290
|
AtGGCT2;2, GGCT2;2, gamma-glutamyl cyclotransferase 2;2 |
-0.201 |
-0.592 |
| 135 |
266649_at |
AT2G25810
|
TIP4;1, tonoplast intrinsic protein 4;1 |
-0.195 |
0.003 |
| 136 |
248486_at |
AT5G51060
|
ATRBOHC, A. THALIANA RESPIRATORY BURST OXIDASE HOMOLOG C, RBOHC, RESPIRATORY BURST OXIDASE HOMOLOG C, RHD2, ROOT HAIR DEFECTIVE 2 |
-0.195 |
8.463 |
| 137 |
247878_at |
AT5G57760
|
unknown |
-0.194 |
4.101 |
| 138 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.192 |
0.225 |
| 139 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.187 |
1.482 |
| 140 |
255007_at |
AT4G10020
|
AtHSD5, hydroxysteroid dehydrogenase 5, HSD5, hydroxysteroid dehydrogenase 5 |
-0.184 |
-0.588 |
| 141 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
-0.184 |
0.953 |
| 142 |
250992_at |
AT5G02260
|
ATEXP9, ATEXPA9, expansin A9, ATHEXP ALPHA 1.10, EXP9, EXPA9, expansin A9 |
-0.180 |
0.405 |
| 143 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
-0.180 |
-1.050 |
| 144 |
266581_at |
AT2G46140
|
[Late embryogenesis abundant protein] |
-0.176 |
4.097 |
| 145 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
-0.172 |
0.782 |
| 146 |
263373_at |
AT2G20515
|
unknown |
-0.171 |
0.139 |
| 147 |
248257_at |
AT5G53410
|
unknown |
-0.170 |
0.225 |
| 148 |
253434_at |
AT4G32500
|
AKT5, K+ transporter 5, KT5, K+ transporter 5 |
-0.168 |
0.231 |
| 149 |
260846_at |
AT1G17300
|
unknown |
-0.167 |
0.225 |
| 150 |
246390_at |
AT1G77330
|
ACO5, ACC oxidase 5 |
-0.166 |
-0.692 |
| 151 |
255805_at |
AT4G10240
|
bbx23, B-box domain protein 23 |
-0.164 |
0.225 |
| 152 |
260181_at |
AT1G70710
|
ATGH9B1, glycosyl hydrolase 9B1, CEL1, CELLULASE 1, GH9B1, glycosyl hydrolase 9B1 |
-0.162 |
-0.243 |
| 153 |
264658_at |
AT1G09910
|
[Rhamnogalacturonate lyase family protein] |
-0.161 |
-0.505 |
| 154 |
249824_at |
AT5G23380
|
unknown |
-0.160 |
0.449 |
| 155 |
256970_at |
AT3G21090
|
ABCG15, ATP-binding cassette G15 |
-0.160 |
0.137 |
| 156 |
251289_at |
AT3G61830
|
ARF18, auxin response factor 18 |
-0.155 |
-1.031 |
| 157 |
250769_at |
AT5G05680
|
EMB2789, EMBRYO DEFECTIVE 2789, MOS7, MODIFIER OF SNC1,7 |
-0.155 |
-0.415 |
| 158 |
251373_at |
AT3G60530
|
GATA4, GATA transcription factor 4 |
-0.152 |
-0.478 |
| 159 |
263738_at |
AT1G60060
|
[Serine/threonine-protein kinase WNK (With No Lysine)-related] |
-0.150 |
-0.258 |
| 160 |
246436_at |
AT5G17440
|
[LUC7 related protein] |
-0.147 |
0.095 |
| 161 |
266125_at |
AT2G45050
|
GATA2, GATA transcription factor 2 |
-0.147 |
0.140 |
| 162 |
252872_at |
AT4G40010
|
SNRK2-7, SUCROSE NONFERMENTING 1-RELATED PROTEIN KINASE 2-7, SNRK2.7, SNF1-related protein kinase 2.7, SRK2F |
-0.146 |
-0.221 |
| 163 |
265274_at |
AT2G28450
|
[zinc finger (CCCH-type) family protein] |
-0.144 |
-0.798 |
| 164 |
256712_at |
AT2G34020
|
[Calcium-binding EF-hand family protein] |
-0.142 |
-0.442 |
| 165 |
258648_at |
AT3G07900
|
[O-fucosyltransferase family protein] |
-0.140 |
-0.733 |
| 166 |
249454_at |
AT5G39520
|
unknown |
-0.138 |
-0.794 |
| 167 |
251039_at |
AT5G02020
|
SIS, Salt Induced Serine rich |
-0.138 |
0.950 |
| 168 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.138 |
0.858 |
| 169 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.136 |
1.047 |
| 170 |
253810_at |
AT4G28220
|
NDB1, NAD(P)H dehydrogenase B1 |
-0.135 |
1.299 |
| 171 |
267075_at |
AT2G41070
|
ATBZIP12, DPBF4, EEL, ENHANCED EM LEVEL |
-0.134 |
0.742 |
| 172 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.134 |
1.481 |
| 173 |
248643_at |
AT5G49130
|
[MATE efflux family protein] |
-0.132 |
0.225 |
| 174 |
245096_at |
AT2G40880
|
ATCYSA, cystatin A, CYSA, cystatin A, FL3-27 |
-0.132 |
0.483 |
| 175 |
251100_at |
AT5G01670
|
[NAD(P)-linked oxidoreductase superfamily protein] |
-0.130 |
0.233 |
| 176 |
261603_at |
AT1G49600
|
ATRBP47A, RNA-binding protein 47A, RBP47A, RNA-binding protein 47A |
-0.130 |
-0.368 |
| 177 |
264524_at |
AT1G10070
|
ATBCAT-2, branched-chain amino acid transaminase 2, BCAT-2, branched-chain amino acid transaminase 2, BCAT2, branched-chain amino acid transferase 2 |
-0.129 |
2.354 |
| 178 |
249164_at |
AT5G42820
|
ATU2AF35B, U2AF35B |
-0.126 |
0.214 |
| 179 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
-0.123 |
0.225 |
| 180 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
-0.123 |
-0.658 |
| 181 |
251870_at |
AT3G54510
|
[Early-responsive to dehydration stress protein (ERD4)] |
-0.123 |
-1.015 |
| 182 |
252740_at |
AT3G43270
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.122 |
-0.165 |
| 183 |
253036_at |
AT4G38340
|
NLP3, NIN-like protein 3 |
-0.121 |
-1.637 |
| 184 |
252098_at |
AT3G51330
|
[Eukaryotic aspartyl protease family protein] |
-0.121 |
1.424 |
| 185 |
262446_at |
AT1G49310
|
unknown |
-0.119 |
0.388 |
| 186 |
266910_at |
AT2G45920
|
[U-box domain-containing protein] |
-0.119 |
0.894 |
| 187 |
266004_at |
AT2G37330
|
ALS3, ALUMINUM SENSITIVE 3 |
-0.119 |
-0.759 |
| 188 |
264902_at |
AT1G23060
|
MDP40, MICROTUBULE DESTABILIZING PROTEIN 40 |
-0.119 |
1.064 |
| 189 |
252446_at |
AT3G47010
|
[Glycosyl hydrolase family protein] |
-0.119 |
-0.955 |
| 190 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.117 |
0.640 |
| 191 |
248708_at |
AT5G48560
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.116 |
2.384 |
| 192 |
257171_at |
AT3G23760
|
unknown |
-0.116 |
-0.996 |
| 193 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
-0.115 |
0.159 |
| 194 |
262580_at |
AT1G15330
|
[Cystathionine beta-synthase (CBS) protein] |
-0.114 |
0.109 |
| 195 |
256452_at |
AT1G75240
|
AtHB33, homeobox protein 33, HB33, homeobox protein 33, ZHD5, zinc-finger homeodomain 5 |
-0.114 |
0.021 |
| 196 |
245325_at |
AT4G14130
|
XTH15, xyloglucan endotransglucosylase/hydrolase 15, XTR7, xyloglucan endotransglycosylase 7 |
-0.114 |
0.271 |
| 197 |
246565_at |
AT5G15530
|
BCCP2, biotin carboxyl carrier protein 2, CAC1-B |
-0.113 |
-0.163 |
| 198 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.113 |
-0.754 |
| 199 |
261657_at |
AT1G50050
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.113 |
0.225 |
| 200 |
256890_at |
AT3G23830
|
AtGRP4, GR-RBP4, GRP4, glycine-rich RNA-binding protein 4 |
-0.113 |
-0.375 |
| 201 |
256600_at |
AT3G14850
|
TBL41, TRICHOME BIREFRINGENCE-LIKE 41 |
-0.112 |
0.843 |
| 202 |
265276_at |
AT2G28400
|
unknown |
-0.112 |
4.468 |
| 203 |
256071_at |
AT1G13640
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
-0.112 |
0.085 |
| 204 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
-0.110 |
3.743 |
| 205 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-0.110 |
-2.305 |
| 206 |
261673_at |
AT1G18280
|
LTPG3, glycosylphosphatidylinositol-anchored lipid protein transfer 3 |
-0.110 |
0.225 |
| 207 |
261880_at |
AT1G50500
|
ATVPS53, ARABIDOPSIS THALIANA VPS53 HOMOLOG, HIT1, HEAT-INTOLERANT 1, VPS53 |
-0.109 |
0.044 |
| 208 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
-0.109 |
-0.501 |
| 209 |
252542_at |
AT3G45770
|
[Polyketide synthase, enoylreductase family protein] |
-0.109 |
-0.688 |
| 210 |
262373_at |
AT1G73120
|
unknown |
-0.108 |
0.490 |
| 211 |
264777_at |
AT1G08630
|
THA1, threonine aldolase 1 |
-0.108 |
4.937 |
| 212 |
250441_at |
AT5G10540
|
TOP2, thimet metalloendopeptidase 2 |
-0.108 |
-0.551 |
| 213 |
252906_at |
AT4G39640
|
GGT1, gamma-glutamyl transpeptidase 1 |
-0.107 |
3.680 |
| 214 |
246596_at |
AT5G14740
|
BETA CA2, BETA CARBONIC ANHYDRASE 2, CA18, CARBONIC ANHYDRASE 18, CA2, carbonic anhydrase 2 |
-0.106 |
-0.499 |
| 215 |
245821_at |
AT1G26270
|
[Phosphatidylinositol 3- and 4-kinase family protein] |
-0.106 |
1.791 |
| 216 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
-0.106 |
-1.310 |
| 217 |
260664_at |
AT1G19510
|
ATRL5, RAD-like 5, RL5, RAD-like 5, RSM4, RADIALIS-LIKE SANT/MYB 4 |
-0.106 |
0.690 |
| 218 |
261272_at |
AT1G26665
|
[Mediator complex, subunit Med10] |
-0.106 |
0.204 |
| 219 |
254327_at |
AT4G22490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.106 |
0.515 |
| 220 |
261299_at |
AT1G48550
|
[Vacuolar protein sorting-associated protein 26] |
-0.106 |
-0.404 |
| 221 |
262205_at |
AT2G01080
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.105 |
-0.369 |
| 222 |
265245_at |
AT2G43060
|
AtIBH1, IBH1, ILI1 binding bHLH 1 |
-0.105 |
-0.014 |
| 223 |
260502_at |
AT1G47270
|
AtTLP6, tubby like protein 6, TLP6, tubby like protein 6 |
-0.104 |
0.782 |
| 224 |
258745_at |
AT3G05920
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.103 |
0.225 |
| 225 |
253388_at |
AT4G32910
|
unknown |
-0.103 |
-0.298 |
| 226 |
256965_at |
AT3G13450
|
DIN4, DARK INDUCIBLE 4 |
-0.102 |
-0.705 |
| 227 |
248029_at |
AT5G55700
|
BAM4, beta-amylase 4, BMY6, BETA-AMYLASE 6 |
-0.102 |
1.049 |
| 228 |
264887_at |
AT1G23120
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.101 |
-0.136 |
| 229 |
263196_at |
AT1G36070
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.101 |
-0.337 |