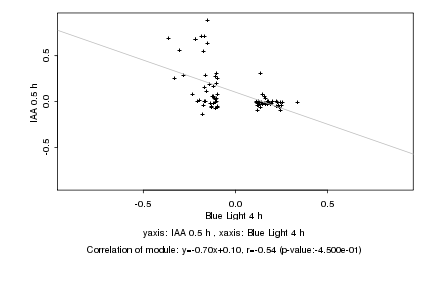
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Blue Light 4 h |
Log2 signal ratio
IAA 0.5 h |
| 1 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
0.335 |
-0.006 |
| 2 |
264931_at |
AT1G60590
|
[Pectin lyase-like superfamily protein] |
0.255 |
-0.004 |
| 3 |
252010_at |
AT3G52740
|
unknown |
0.249 |
-0.035 |
| 4 |
252661_at |
AT3G44450
|
unknown |
0.244 |
-0.087 |
| 5 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.244 |
-0.010 |
| 6 |
245628_at |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
0.238 |
-0.063 |
| 7 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.233 |
-0.038 |
| 8 |
252123_at |
AT3G51240
|
F3'H, F3H, flavanone 3-hydroxylase, TT6, TRANSPARENT TESTA 6 |
0.227 |
-0.005 |
| 9 |
249191_at |
AT5G42760
|
[Leucine carboxyl methyltransferase] |
0.222 |
0.001 |
| 10 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
0.218 |
-0.048 |
| 11 |
257954_at |
AT3G21760
|
HYR1, HYPOSTATIN RESISTANCE 1 |
0.201 |
0.004 |
| 12 |
258321_at |
AT3G22840
|
ELIP, ELIP1, EARLY LIGHT-INDUCABLE PROTEIN |
0.192 |
-0.021 |
| 13 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.188 |
-0.024 |
| 14 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.183 |
-0.020 |
| 15 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
0.181 |
-0.011 |
| 16 |
250794_at |
AT5G05270
|
AtCHIL, CHIL, chalcone isomerase like |
0.178 |
-0.015 |
| 17 |
251020_at |
AT5G02270
|
ABCI20, ATP-binding cassette I20, ATNAP9, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN 9, NAP9, non-intrinsic ABC protein 9 |
0.178 |
-0.012 |
| 18 |
250420_at |
AT5G11260
|
HY5, ELONGATED HYPOCOTYL 5, TED 5, REVERSAL OF THE DET PHENOTYPE 5 |
0.177 |
-0.010 |
| 19 |
256542_at |
AT1G42550
|
PMI1, PLASTID MOVEMENT IMPAIRED1 |
0.177 |
0.004 |
| 20 |
246468_at |
AT5G17050
|
UGT78D2, UDP-glucosyl transferase 78D2 |
0.174 |
0.006 |
| 21 |
259076_at |
AT3G02140
|
AFP4, ABI FIVE BINDING PROTEIN 4, TMAC2, TWO OR MORE ABRES-CONTAINING GENE 2 |
0.174 |
-0.026 |
| 22 |
246597_at |
AT5G14760
|
AO, L-aspartate oxidase, FIN4, FLAGELLIN-INSENSITIVE 4 |
0.158 |
0.033 |
| 23 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.158 |
-0.028 |
| 24 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.157 |
-0.016 |
| 25 |
254634_at |
AT4G18650
|
[transcription factor-related] |
0.153 |
0.063 |
| 26 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
0.146 |
-0.031 |
| 27 |
256097_at |
AT1G13670
|
unknown |
0.144 |
0.085 |
| 28 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.139 |
-0.005 |
| 29 |
263739_at |
AT2G21320
|
BBX18, B-box domain protein 18 |
0.135 |
-0.064 |
| 30 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.133 |
0.316 |
| 31 |
258696_at |
AT3G09650
|
CRM3, HCF152, HIGH CHLOROPHYLL FLUORESCENCE 152 |
0.129 |
-0.002 |
| 32 |
247596_at |
AT5G60840
|
unknown |
0.129 |
-0.026 |
| 33 |
253155_at |
AT4G35720
|
unknown |
0.129 |
-0.006 |
| 34 |
263796_at |
AT2G24540
|
AFR, ATTENUATED FAR-RED RESPONSE |
0.125 |
-0.036 |
| 35 |
262536_at |
AT1G17100
|
AtHBP1, HBP1, haem-binding protein 1 |
0.125 |
-0.001 |
| 36 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
0.125 |
0.008 |
| 37 |
251827_at |
AT3G55120
|
A11, AtCHI, CFI, CHALCONE FLAVANONE ISOMERASE, CHI, chalcone isomerase, TT5, TRANSPARENT TESTA 5 |
0.124 |
-0.021 |
| 38 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
0.121 |
-0.000 |
| 39 |
263787_at |
AT2G46420
|
unknown |
0.117 |
-0.033 |
| 40 |
267070_at |
AT2G41000
|
[Chaperone DnaJ-domain superfamily protein] |
0.116 |
-0.021 |
| 41 |
265634_at |
AT2G25530
|
[AFG1-like ATPase family protein] |
0.115 |
-0.088 |
| 42 |
249406_at |
AT5G40210
|
UMAMIT42, Usually multiple acids move in and out Transporters 42 |
0.113 |
-0.007 |
| 43 |
259537_at |
AT1G12370
|
PHR1, photolyase 1, UVR2, UV RESISTANCE 2 |
0.112 |
0.006 |
| 44 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.366 |
0.694 |
| 45 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.331 |
0.256 |
| 46 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.308 |
0.567 |
| 47 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.284 |
0.291 |
| 48 |
249008_at |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.235 |
0.085 |
| 49 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.218 |
0.680 |
| 50 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.217 |
0.679 |
| 51 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.211 |
0.004 |
| 52 |
247533_at |
AT5G61570
|
[Protein kinase superfamily protein] |
-0.196 |
0.012 |
| 53 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.188 |
0.716 |
| 54 |
255608_at |
AT4G01140
|
unknown |
-0.184 |
-0.131 |
| 55 |
248432_at |
AT5G51390
|
unknown |
-0.178 |
-0.035 |
| 56 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.175 |
0.555 |
| 57 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.172 |
0.712 |
| 58 |
248460_at |
AT5G50915
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.169 |
0.010 |
| 59 |
259274_at |
AT3G01220
|
ATHB20, homeobox protein 20, HB20, homeobox protein 20 |
-0.168 |
0.155 |
| 60 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.167 |
0.286 |
| 61 |
262236_at |
AT1G48330
|
unknown |
-0.163 |
0.002 |
| 62 |
258541_at |
AT3G07000
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.160 |
0.118 |
| 63 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.155 |
0.638 |
| 64 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.154 |
0.894 |
| 65 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.146 |
0.190 |
| 66 |
254954_at |
AT4G10910
|
unknown |
-0.139 |
-0.019 |
| 67 |
251771_at |
AT3G56000
|
ATCSLA14, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A14, CSLA14, cellulose synthase like A14 |
-0.133 |
-0.050 |
| 68 |
264379_at |
AT2G25200
|
unknown |
-0.132 |
-0.055 |
| 69 |
262010_at |
AT1G35612
|
[expressed protein] |
-0.125 |
0.055 |
| 70 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
-0.124 |
0.065 |
| 71 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.122 |
0.164 |
| 72 |
263876_at |
AT2G21880
|
ATRAB7A, RAB GTPase homolog 7A, ATRABG2, ARABIDOPSIS RAB GTPASE HOMOLOG G2, RAB7A, RAB GTPase homolog 7A |
-0.120 |
-0.017 |
| 73 |
246350_at |
AT1G16650
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.118 |
0.040 |
| 74 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.113 |
-0.076 |
| 75 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
-0.112 |
0.023 |
| 76 |
261158_at |
AT1G34500
|
[MBOAT (membrane bound O-acyl transferase) family protein] |
-0.112 |
-0.006 |
| 77 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.111 |
0.279 |
| 78 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.106 |
0.199 |
| 79 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.104 |
0.313 |
| 80 |
245794_at |
AT1G32170
|
XTH30, xyloglucan endotransglucosylase/hydrolase 30, XTR4, xyloglucan endotransglycosylase 4 |
-0.103 |
0.011 |
| 81 |
250616_at |
AT5G07280
|
EMS1, EXCESS MICROSPOROCYTES1, EXS, EXTRA SPOROGENOUS CELLS |
-0.103 |
0.041 |
| 82 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.102 |
0.253 |
| 83 |
266656_at |
AT2G25900
|
ATCTH, ATTZF1, A. THALIANA TANDEM ZINC FINGER PROTEIN 1 |
-0.100 |
-0.048 |
| 84 |
248130_at |
AT5G54790
|
unknown |
-0.100 |
0.083 |
| 85 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.100 |
-0.055 |