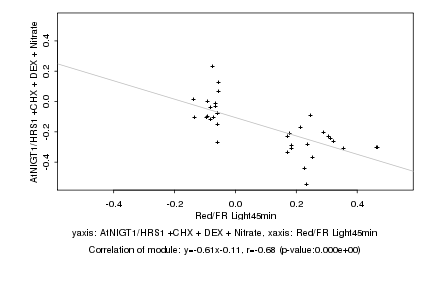
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Red/FR Light45min |
Log2 signal ratio
AtNIGT1/HRS1 +CHX + DEX + Nitrate |
| 1 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.464 |
-0.300 |
| 2 |
263374_at |
AT2G20560
|
[DNAJ heat shock family protein] |
0.463 |
-0.299 |
| 3 |
258133_at |
AT3G24500
|
ATMBF1C, MBF1C, multiprotein bridging factor 1C |
0.353 |
-0.306 |
| 4 |
260248_at |
AT1G74310
|
ATHSP101, heat shock protein 101, HOT1, HSP101, heat shock protein 101 |
0.321 |
-0.258 |
| 5 |
248332_at |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
0.310 |
-0.242 |
| 6 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
0.303 |
-0.227 |
| 7 |
256245_at |
AT3G12580
|
ATHSP70, ARABIDOPSIS HEAT SHOCK PROTEIN 70, HSP70, heat shock protein 70 |
0.288 |
-0.203 |
| 8 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.250 |
-0.364 |
| 9 |
261081_at |
AT1G07350
|
SR45a, serine/arginine rich-like protein 45a |
0.246 |
-0.091 |
| 10 |
248434_at |
AT5G51440
|
[HSP20-like chaperones superfamily protein] |
0.234 |
-0.277 |
| 11 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.231 |
-0.541 |
| 12 |
256518_at |
AT1G66080
|
unknown |
0.225 |
-0.436 |
| 13 |
256999_at |
AT3G14200
|
[Chaperone DnaJ-domain superfamily protein] |
0.212 |
-0.171 |
| 14 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.183 |
-0.307 |
| 15 |
264402_at |
AT2G25140
|
CLPB-M, CASEIN LYTIC PROTEINASE B-M, CLPB4, casein lytic proteinase B4, HSP98.7, HEAT SHOCK PROTEIN 98.7 |
0.181 |
-0.285 |
| 16 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.177 |
-0.205 |
| 17 |
248045_at |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
0.170 |
-0.225 |
| 18 |
254211_at |
AT4G23570
|
SGT1A |
0.168 |
-0.330 |
| 19 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
-0.141 |
0.014 |
| 20 |
267294_at |
AT2G23670
|
YCF37, homolog of Synechocystis YCF37 |
-0.136 |
-0.099 |
| 21 |
246983_at |
AT5G67200
|
[Leucine-rich repeat protein kinase family protein] |
-0.096 |
-0.100 |
| 22 |
252268_at |
AT3G49650
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.094 |
0.005 |
| 23 |
249572_at |
AT5G37630
|
EMB2656, EMBRYO DEFECTIVE 2656 |
-0.093 |
-0.096 |
| 24 |
266862_at |
AT2G26950
|
AtMYB104, myb domain protein 104, MYB104, myb domain protein 104 |
-0.085 |
-0.116 |
| 25 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.082 |
-0.038 |
| 26 |
265297_at |
AT2G14080
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.078 |
0.232 |
| 27 |
250567_at |
AT5G08090
|
unknown |
-0.074 |
-0.104 |
| 28 |
263859_at |
AT2G04360
|
unknown |
-0.067 |
-0.030 |
| 29 |
254808_at |
AT4G12740
|
[HhH-GPD base excision DNA repair family protein] |
-0.066 |
-0.008 |
| 30 |
246258_at |
AT1G31840
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.062 |
-0.077 |
| 31 |
267298_at |
AT2G23760
|
BLH4, BEL1-like homeodomain 4, SAW2, SAWTOOTH 2 |
-0.061 |
-0.268 |
| 32 |
260316_at |
AT1G63810
|
unknown |
-0.060 |
-0.073 |
| 33 |
264692_at |
AT1G70000
|
[myb-like transcription factor family protein] |
-0.059 |
-0.149 |
| 34 |
253541_at |
AT4G31630
|
[Transcriptional factor B3 family protein] |
-0.058 |
0.131 |
| 35 |
246615_at |
AT5G35420
|
unknown |
-0.056 |
0.072 |