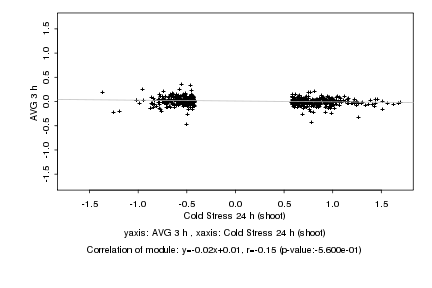
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Cold Stress 24 h (shoot) |
Log2 signal ratio
AVG 3 h |
| 1 |
245749_at |
AT1G51090
|
[Heavy metal transport/detoxification superfamily protein ] |
1.693 |
-0.020 |
| 2 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
1.671 |
-0.023 |
| 3 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
1.622 |
-0.059 |
| 4 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
1.556 |
-0.026 |
| 5 |
256114_at |
AT1G16850
|
unknown |
1.509 |
0.009 |
| 6 |
253416_at |
AT4G33070
|
AtPDC1, PDC1, pyruvate decarboxylase 1 |
1.504 |
-0.147 |
| 7 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
1.460 |
0.045 |
| 8 |
266141_at |
AT2G02120
|
LCR70, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 70, PDF2.1 |
1.439 |
0.057 |
| 9 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
1.437 |
-0.032 |
| 10 |
267036_at |
AT2G38465
|
unknown |
1.425 |
-0.088 |
| 11 |
253595_at |
AT4G30830
|
unknown |
1.404 |
-0.055 |
| 12 |
263495_at |
AT2G42530
|
COR15B, cold regulated 15b |
1.379 |
0.039 |
| 13 |
256221_at |
AT1G56300
|
[Chaperone DnaJ-domain superfamily protein] |
1.363 |
-0.053 |
| 14 |
262452_at |
AT1G11210
|
unknown |
1.329 |
-0.069 |
| 15 |
258347_at |
AT3G17520
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.303 |
-0.007 |
| 16 |
256017_at |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
1.296 |
-0.024 |
| 17 |
253322_at |
AT4G33980
|
unknown |
1.264 |
-0.035 |
| 18 |
245319_at |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
1.264 |
-0.314 |
| 19 |
264617_at |
AT2G17660
|
[RPM1-interacting protein 4 (RIN4) family protein] |
1.245 |
-0.018 |
| 20 |
262307_at |
AT1G71000
|
[Chaperone DnaJ-domain superfamily protein] |
1.242 |
-0.082 |
| 21 |
257154_at |
AT3G27210
|
unknown |
1.240 |
0.008 |
| 22 |
251793_at |
AT3G55580
|
[Regulator of chromosome condensation (RCC1) family protein] |
1.224 |
0.059 |
| 23 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
1.220 |
-0.039 |
| 24 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
1.204 |
-0.003 |
| 25 |
258805_at |
AT3G04010
|
[O-Glycosyl hydrolases family 17 protein] |
1.202 |
-0.036 |
| 26 |
249741_at |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
1.192 |
-0.030 |
| 27 |
254066_at |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
1.160 |
0.071 |
| 28 |
263789_at |
AT2G24560
|
[GDSL-like Lipase/Acylhydrolase family protein] |
1.159 |
0.018 |
| 29 |
261077_at |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
1.156 |
-0.032 |
| 30 |
251336_at |
AT3G61190
|
BAP1, BON association protein 1 |
1.147 |
0.021 |
| 31 |
254818_at |
AT4G12470
|
AZI1, azelaic acid induced 1 |
1.147 |
-0.040 |
| 32 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
1.144 |
0.056 |
| 33 |
266532_at |
AT2G16890
|
[UDP-Glycosyltransferase superfamily protein] |
1.130 |
0.017 |
| 34 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
1.119 |
0.116 |
| 35 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
1.110 |
0.040 |
| 36 |
261613_at |
AT1G49720
|
ABF1, abscisic acid responsive element-binding factor 1, AtABF1 |
1.082 |
-0.003 |
| 37 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
1.078 |
0.079 |
| 38 |
245711_at |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
1.073 |
0.036 |
| 39 |
250826_at |
AT5G05220
|
unknown |
1.073 |
0.004 |
| 40 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
1.069 |
-0.001 |
| 41 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
1.062 |
0.102 |
| 42 |
260556_at |
AT2G43620
|
[Chitinase family protein] |
1.060 |
-0.064 |
| 43 |
257280_at |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
1.053 |
0.009 |
| 44 |
265276_at |
AT2G28400
|
unknown |
1.042 |
0.107 |
| 45 |
248337_at |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
1.038 |
-0.043 |
| 46 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
1.037 |
0.022 |
| 47 |
245807_at |
AT1G46768
|
RAP2.1, related to AP2 1 |
1.036 |
0.021 |
| 48 |
246831_at |
AT5G26340
|
ATSTP13, SUGAR TRANSPORT PROTEIN 13, MSS1, STP13, SUGAR TRANSPORT PROTEIN 13 |
1.030 |
0.097 |
| 49 |
259442_at |
AT1G02310
|
MAN1, endo-beta-mannanase 1 |
1.026 |
-0.010 |
| 50 |
248959_at |
AT5G45630
|
unknown |
1.016 |
-0.136 |
| 51 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
1.014 |
0.001 |
| 52 |
251753_at |
AT3G55760
|
unknown |
1.008 |
-0.051 |
| 53 |
259977_at |
AT1G76590
|
[PLATZ transcription factor family protein] |
1.005 |
0.008 |
| 54 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
1.005 |
-0.070 |
| 55 |
245904_at |
AT5G11110
|
ATSPS2F, sucrose phosphate synthase 2F, KNS2, KAONASHI 2, SPS1, SUCROSE PHOSPHATE SYNTHASE 1, SPS2F, sucrose phosphate synthase 2F, SPSA2, sucrose-phosphate synthase A2 |
1.001 |
0.051 |
| 56 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.994 |
-0.005 |
| 57 |
248744_at |
AT5G48250
|
BBX8, B-box domain protein 8 |
0.992 |
0.066 |
| 58 |
255331_at |
AT4G04330
|
AtRbcX1, RbcX1, homologue of cyanobacterial RbcX 1 |
0.992 |
-0.019 |
| 59 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.991 |
0.041 |
| 60 |
250062_at |
AT5G17760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.982 |
-0.059 |
| 61 |
263931_at |
AT2G36220
|
unknown |
0.982 |
-0.228 |
| 62 |
267411_at |
AT2G34930
|
[disease resistance family protein / LRR family protein] |
0.981 |
-0.101 |
| 63 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.980 |
0.076 |
| 64 |
246922_at |
AT5G25110
|
CIPK25, CBL-interacting protein kinase 25, SnRK3.25, SNF1-RELATED PROTEIN KINASE 3.25 |
0.980 |
-0.085 |
| 65 |
262050_at |
AT1G80130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.977 |
-0.140 |
| 66 |
261248_at |
AT1G20030
|
[Pathogenesis-related thaumatin superfamily protein] |
0.974 |
0.028 |
| 67 |
253104_at |
AT4G36010
|
[Pathogenesis-related thaumatin superfamily protein] |
0.968 |
-0.056 |
| 68 |
254075_at |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
0.967 |
0.011 |
| 69 |
264217_at |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
0.965 |
0.096 |
| 70 |
264580_at |
AT1G05340
|
unknown |
0.964 |
0.076 |
| 71 |
260688_at |
AT1G17665
|
unknown |
0.963 |
-0.099 |
| 72 |
254210_at |
AT4G23450
|
AIRP1, ABA Insensitive RING Protein 1, AtAIRP1 |
0.958 |
-0.019 |
| 73 |
246435_at |
AT5G17460
|
unknown |
0.958 |
0.025 |
| 74 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
0.956 |
0.016 |
| 75 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.956 |
0.091 |
| 76 |
264019_at |
AT2G21130
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
0.955 |
-0.004 |
| 77 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.954 |
0.010 |
| 78 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.954 |
-0.133 |
| 79 |
253872_at |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
0.951 |
-0.011 |
| 80 |
255607_at |
AT4G01130
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.945 |
0.121 |
| 81 |
252908_at |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
0.943 |
-0.038 |
| 82 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.941 |
0.013 |
| 83 |
261187_at |
AT1G32860
|
[Glycosyl hydrolase superfamily protein] |
0.935 |
0.023 |
| 84 |
247937_at |
AT5G57110
|
ACA8, autoinhibited Ca2+ -ATPase, isoform 8, AT-ACA8, AUTOINHIBITED CA2+ -ATPASE, ISOFORM 8 |
0.934 |
0.057 |
| 85 |
261318_at |
AT1G53035
|
unknown |
0.932 |
-0.093 |
| 86 |
245699_at |
AT5G04250
|
[Cysteine proteinases superfamily protein] |
0.932 |
-0.008 |
| 87 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.930 |
0.006 |
| 88 |
248410_at |
AT5G51570
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.929 |
-0.016 |
| 89 |
254074_at |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
0.929 |
0.017 |
| 90 |
263548_at |
AT2G21660
|
ATGRP7, GLYCINE RICH PROTEIN 7, CCR2, cold, circadian rhythm, and rna binding 2, GR-RBP7, GLYCINE-RICH RNA-BINDING PROTEIN 7, GRP7, GLYCINE-RICH RNA-BINDING PROTEIN 7 |
0.929 |
-0.032 |
| 91 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.927 |
0.027 |
| 92 |
258893_at |
AT3G05660
|
AtRLP33, receptor like protein 33, RLP33, receptor like protein 33 |
0.925 |
-0.109 |
| 93 |
267461_at |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
0.924 |
-0.005 |
| 94 |
252956_at |
AT4G38580
|
ATFP6, farnesylated protein 6, FP6, farnesylated protein 6, HIPP26, HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN 26 |
0.923 |
-0.056 |
| 95 |
248236_at |
AT5G53870
|
AtENODL1, ENODL1, early nodulin-like protein 1 |
0.919 |
-0.207 |
| 96 |
267280_at |
AT2G19450
|
ABX45, AS11, ATDGAT, Arabidopsis thaliana acyl-CoA:diacylglycerol acyltransferase, AtDGAT1, DGAT1, acyl-CoA:diacylglycerol acyltransferase 1, RDS1, TAG1, TRIACYLGLYCEROL BIOSYNTHESIS DEFECT 1 |
0.917 |
-0.008 |
| 97 |
245627_at |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
0.915 |
0.071 |
| 98 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.915 |
0.008 |
| 99 |
264000_at |
AT2G22500
|
ATPUMP5, PLANT UNCOUPLING MITOCHONDRIAL PROTEIN 5, DIC1, DICARBOXYLATE CARRIER 1, UCP5, uncoupling protein 5 |
0.910 |
-0.029 |
| 100 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.907 |
0.053 |
| 101 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.905 |
0.090 |
| 102 |
258139_at |
AT3G24520
|
AT-HSFC1, HSFC1, heat shock transcription factor C1 |
0.899 |
0.012 |
| 103 |
257226_at |
AT3G27880
|
unknown |
0.899 |
0.052 |
| 104 |
267080_at |
AT2G41190
|
[Transmembrane amino acid transporter family protein] |
0.897 |
0.026 |
| 105 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.895 |
0.055 |
| 106 |
246001_at |
AT5G20790
|
unknown |
0.894 |
0.089 |
| 107 |
253293_at |
AT4G33905
|
[Peroxisomal membrane 22 kDa (Mpv17/PMP22) family protein] |
0.892 |
0.045 |
| 108 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
0.880 |
0.140 |
| 109 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.878 |
0.093 |
| 110 |
262892_at |
AT1G79440
|
ALDH5F1, aldehyde dehydrogenase 5F1, ENF1, ENLARGED FIL EXPRESSION DOMAIN 1, SSADH, SUCCINIC SEMIALDEHYDE DEHYDROGENASE, SSADH1, SUCCINIC SEMIALDEHYDE DEHYDROGENASE 1 |
0.873 |
-0.006 |
| 111 |
261610_at |
AT1G49560
|
[Homeodomain-like superfamily protein] |
0.870 |
-0.056 |
| 112 |
245635_at |
AT1G25250
|
AtIDD16, indeterminate(ID)-domain 16, IDD16, indeterminate(ID)-domain 16 |
0.870 |
-0.035 |
| 113 |
255479_at |
AT4G02380
|
AtLEA5, Arabidopsis thaliana late embryogenensis abundant like 5, SAG21, senescence-associated gene 21 |
0.867 |
-0.059 |
| 114 |
251494_at |
AT3G59350
|
[Protein kinase superfamily protein] |
0.865 |
0.003 |
| 115 |
258397_at |
AT3G15357
|
unknown |
0.863 |
-0.048 |
| 116 |
263352_at |
AT2G22080
|
unknown |
0.863 |
-0.016 |
| 117 |
260264_at |
AT1G68500
|
unknown |
0.863 |
-0.016 |
| 118 |
247177_at |
AT5G65300
|
unknown |
0.863 |
-0.097 |
| 119 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.862 |
0.044 |
| 120 |
252944_at |
AT4G39320
|
[microtubule-associated protein-related] |
0.861 |
-0.000 |
| 121 |
250781_at |
AT5G05410
|
DREB2, DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN 2, DREB2A, DRE-binding protein 2A |
0.861 |
0.044 |
| 122 |
262881_at |
AT1G64890
|
[Major facilitator superfamily protein] |
0.861 |
0.069 |
| 123 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.860 |
0.067 |
| 124 |
263497_at |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
0.858 |
0.012 |
| 125 |
250412_at |
AT5G11150
|
ATVAMP713, vesicle-associated membrane protein 713, VAMP713, VAMP713, vesicle-associated membrane protein 713 |
0.858 |
-0.039 |
| 126 |
257876_at |
AT3G17130
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.856 |
-0.000 |
| 127 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
0.851 |
-0.111 |
| 128 |
250556_at |
AT5G07920
|
ATDGK1, DIACYLGLYCEROL KINASE 1, DGK1, diacylglycerol kinase1 |
0.842 |
0.062 |
| 129 |
250158_at |
AT5G15190
|
unknown |
0.839 |
0.032 |
| 130 |
247525_at |
AT5G61380
|
APRR1, AtTOC1, TIMING OF CAB EXPRESSION 1, PRR1, PSEUDO-RESPONSE REGULATOR 1, TOC1, TIMING OF CAB EXPRESSION 1 |
0.839 |
-0.036 |
| 131 |
266503_at |
AT2G47780
|
[Rubber elongation factor protein (REF)] |
0.834 |
-0.062 |
| 132 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
0.830 |
-0.111 |
| 133 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.829 |
0.060 |
| 134 |
265354_at |
AT2G16700
|
ADF5, actin depolymerizing factor 5, ATADF5 |
0.827 |
0.039 |
| 135 |
246432_at |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
0.826 |
-0.019 |
| 136 |
255494_at |
AT4G02710
|
NET1C, Networked 1C |
0.824 |
-0.010 |
| 137 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.824 |
-0.028 |
| 138 |
250053_at |
AT5G17850
|
[Sodium/calcium exchanger family protein] |
0.822 |
-0.019 |
| 139 |
257593_at |
AT3G24840
|
[Sec14p-like phosphatidylinositol transfer family protein] |
0.822 |
0.036 |
| 140 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
0.815 |
0.033 |
| 141 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.812 |
-0.051 |
| 142 |
260410_at |
AT1G69870
|
AtNPF2.13, NPF2.13, NRT1/ PTR family 2.13, NRT1.7, nitrate transporter 1.7 |
0.808 |
-0.044 |
| 143 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
0.807 |
-0.050 |
| 144 |
259705_at |
AT1G77450
|
anac032, NAC domain containing protein 32, NAC032, NAC domain containing protein 32 |
0.806 |
0.217 |
| 145 |
248505_at |
AT5G50360
|
unknown |
0.804 |
0.037 |
| 146 |
245998_at |
AT5G20830
|
ASUS1, atsus1, SUS1, sucrose synthase 1 |
0.802 |
-0.084 |
| 147 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
0.799 |
-0.216 |
| 148 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.798 |
0.035 |
| 149 |
263221_at |
AT1G30620
|
HSR8, HIGH SUGAR RESPONSE8, MUR4, MURUS 4, UXE1, UDP-D-XYLOSE 4-EPIMERASE 1 |
0.797 |
0.040 |
| 150 |
264246_at |
AT1G60140
|
ATTPS10, trehalose phosphate synthase, TPS10, trehalose phosphate synthase, TPS10, TREHALOSE PHOSPHATE SYNTHASE 10 |
0.796 |
-0.081 |
| 151 |
260357_at |
AT1G69260
|
AFP1, ABI five binding protein |
0.792 |
-0.048 |
| 152 |
266514_at |
AT2G47890
|
[B-box type zinc finger protein with CCT domain] |
0.792 |
0.035 |
| 153 |
259479_at |
AT1G19020
|
unknown |
0.789 |
-0.028 |
| 154 |
246419_at |
AT5G17030
|
UGT78D3, UDP-glucosyl transferase 78D3 |
0.787 |
0.009 |
| 155 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.786 |
0.004 |
| 156 |
259516_at |
AT1G20450
|
ERD10, EARLY RESPONSIVE TO DEHYDRATION 10, LTI29, LOW TEMPERATURE INDUCED 29, LTI45, LOW TEMPERATURE INDUCED 45 |
0.785 |
-0.044 |
| 157 |
261263_at |
AT1G26790
|
[Dof-type zinc finger DNA-binding family protein] |
0.784 |
0.017 |
| 158 |
262496_at |
AT1G21790
|
[TRAM, LAG1 and CLN8 (TLC) lipid-sensing domain containing protein] |
0.783 |
0.042 |
| 159 |
249456_at |
AT5G39410
|
[Saccharopine dehydrogenase ] |
0.778 |
-0.003 |
| 160 |
252321_at |
AT3G48510
|
unknown |
0.775 |
0.015 |
| 161 |
251603_at |
AT3G57760
|
[Protein kinase superfamily protein] |
0.775 |
0.012 |
| 162 |
251927_at |
AT3G53990
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
0.775 |
0.017 |
| 163 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.773 |
-0.002 |
| 164 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.773 |
-0.430 |
| 165 |
261726_at |
AT1G76270
|
[O-fucosyltransferase family protein] |
0.773 |
-0.100 |
| 166 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.771 |
0.003 |
| 167 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.767 |
-0.197 |
| 168 |
258078_at |
AT3G25870
|
unknown |
0.765 |
0.193 |
| 169 |
247776_at |
AT5G58700
|
ATPLC4, PLC4, phosphatidylinositol-speciwc phospholipase C4 |
0.765 |
-0.018 |
| 170 |
251768_at |
AT3G55940
|
[Phosphoinositide-specific phospholipase C family protein] |
0.763 |
-0.001 |
| 171 |
247786_at |
AT5G58600
|
PMR5, POWDERY MILDEW RESISTANT 5, TBL44, TRICHOME BIREFRINGENCE-LIKE 44 |
0.761 |
-0.160 |
| 172 |
246779_at |
AT5G27520
|
AtPNC2, PNC2, peroxisomal adenine nucleotide carrier 2 |
0.753 |
-0.053 |
| 173 |
264314_at |
AT1G70420
|
unknown |
0.751 |
0.067 |
| 174 |
259417_at |
AT1G02340
|
FBI1, HFR1, LONG HYPOCOTYL IN FAR-RED, REP1, REDUCED PHYTOCHROME SIGNALING 1, RSF1, REDUCED SENSITIVITY TO FAR-RED LIGHT 1 |
0.749 |
0.108 |
| 175 |
247655_at |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
0.746 |
0.196 |
| 176 |
261937_at |
AT1G22570
|
[Major facilitator superfamily protein] |
0.744 |
0.072 |
| 177 |
248820_at |
AT5G47060
|
unknown |
0.744 |
-0.006 |
| 178 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.740 |
0.026 |
| 179 |
260744_at |
AT1G15010
|
unknown |
0.737 |
0.040 |
| 180 |
265093_at |
AT1G03905
|
ABCI19, ATP-binding cassette I19 |
0.737 |
-0.102 |
| 181 |
262477_at |
AT1G11220
|
unknown |
0.736 |
-0.031 |
| 182 |
264045_at |
AT2G22450
|
AtRIBA2, RIBA2, homolog of ribA 2 |
0.735 |
-0.012 |
| 183 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
0.733 |
-0.054 |
| 184 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.731 |
-0.001 |
| 185 |
261745_at |
AT1G08500
|
AtENODL18, ENODL18, early nodulin-like protein 18 |
0.730 |
0.111 |
| 186 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
0.727 |
0.062 |
| 187 |
254188_at |
AT4G23920
|
ATUGE2, UDP-GLC 4-EPIMERASE 2, UGE2, UDP-D-glucose/UDP-D-galactose 4-epimerase 2 |
0.724 |
0.001 |
| 188 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
0.723 |
-0.121 |
| 189 |
251620_at |
AT3G58060
|
[Cation efflux family protein] |
0.719 |
-0.008 |
| 190 |
262296_at |
AT1G27630
|
CYCT1;3, cyclin T 1;3 |
0.717 |
0.036 |
| 191 |
264989_at |
AT1G27200
|
unknown |
0.716 |
-0.023 |
| 192 |
267631_at |
AT2G42150
|
[DNA-binding bromodomain-containing protein] |
0.713 |
0.061 |
| 193 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
0.713 |
0.092 |
| 194 |
265886_at |
AT2G25620
|
AtDBP1, DNA-binding protein phosphatase 1, DBP1, DNA-binding protein phosphatase 1 |
0.710 |
-0.045 |
| 195 |
254580_at |
AT4G19390
|
[Uncharacterised protein family (UPF0114)] |
0.709 |
0.013 |
| 196 |
265290_at |
AT2G22590
|
[UDP-Glycosyltransferase superfamily protein] |
0.709 |
0.031 |
| 197 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.707 |
0.016 |
| 198 |
259137_at |
AT3G10300
|
[Calcium-binding EF-hand family protein] |
0.706 |
-0.007 |
| 199 |
263379_at |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
0.706 |
0.010 |
| 200 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.705 |
-0.011 |
| 201 |
248799_at |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
0.702 |
-0.100 |
| 202 |
252573_at |
AT3G45260
|
[C2H2-like zinc finger protein] |
0.699 |
-0.027 |
| 203 |
259570_at |
AT1G20440
|
AtCOR47, COR47, cold-regulated 47, RD17 |
0.696 |
0.044 |
| 204 |
259244_at |
AT3G07650
|
BBX7, B-box domain protein 7, COL9, CONSTANS-like 9 |
0.692 |
0.012 |
| 205 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.690 |
-0.126 |
| 206 |
256296_at |
AT1G69480
|
[EXS (ERD1/XPR1/SYG1) family protein] |
0.690 |
0.071 |
| 207 |
253818_at |
AT4G28330
|
unknown |
0.689 |
-0.066 |
| 208 |
259173_at |
AT3G03640
|
BGLU25, beta glucosidase 25, GLUC |
0.687 |
-0.006 |
| 209 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.685 |
-0.266 |
| 210 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.684 |
0.040 |
| 211 |
252557_at |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
0.684 |
0.019 |
| 212 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.683 |
-0.077 |
| 213 |
266119_at |
AT2G02100
|
LCR69, low-molecular-weight cysteine-rich 69, PDF2.2 |
0.682 |
-0.023 |
| 214 |
253809_at |
AT4G28320
|
AtMAN5, MAN5, endo-beta-mannase 5 |
0.680 |
-0.022 |
| 215 |
249619_at |
AT5G37500
|
GORK, gated outwardly-rectifying K+ channel |
0.678 |
-0.097 |
| 216 |
259971_at |
AT1G76580
|
[Squamosa promoter-binding protein-like (SBP domain) transcription factor family protein] |
0.678 |
-0.016 |
| 217 |
260674_at |
AT1G19370
|
unknown |
0.677 |
-0.037 |
| 218 |
250194_at |
AT5G14550
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.675 |
0.101 |
| 219 |
264458_at |
AT1G10410
|
unknown |
0.675 |
-0.075 |
| 220 |
249204_at |
AT5G42570
|
[B-cell receptor-associated 31-like] |
0.673 |
-0.019 |
| 221 |
246796_at |
AT5G26770
|
unknown |
0.673 |
-0.014 |
| 222 |
253215_at |
AT4G34950
|
[Major facilitator superfamily protein] |
0.673 |
0.057 |
| 223 |
248138_at |
AT5G54960
|
PDC2, pyruvate decarboxylase-2 |
0.672 |
-0.024 |
| 224 |
254304_at |
AT4G22270
|
ATMRB1, ARABIDOPSIS MEMBRANE RELATED BIGGER1, MRB1, MEMBRANE RELATED BIGGER1 |
0.671 |
0.088 |
| 225 |
247498_at |
AT5G61810
|
APC1, ATP/phosphate carrier 1 |
0.670 |
0.055 |
| 226 |
252368_at |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
0.668 |
-0.006 |
| 227 |
267576_at |
AT2G30640
|
MUG2, MUSTANG 2 |
0.668 |
0.041 |
| 228 |
257237_at |
AT3G14890
|
[phosphoesterase] |
0.663 |
-0.100 |
| 229 |
259595_at |
AT1G28050
|
BBX13, B-box domain protein 13 |
0.663 |
-0.015 |
| 230 |
260489_at |
AT1G51610
|
[Cation efflux family protein] |
0.662 |
0.057 |
| 231 |
258827_at |
AT3G07150
|
unknown |
0.661 |
-0.012 |
| 232 |
262760_at |
AT1G10770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.659 |
-0.089 |
| 233 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
0.655 |
0.144 |
| 234 |
250987_at |
AT5G02860
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.653 |
-0.006 |
| 235 |
253581_at |
AT4G30660
|
[Low temperature and salt responsive protein family] |
0.653 |
0.000 |
| 236 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.653 |
-0.062 |
| 237 |
264972_at |
AT1G67370
|
ASY1, ASYNAPTIC 1, ATASY1 |
0.652 |
0.012 |
| 238 |
261168_at |
AT1G04945
|
[HIT-type Zinc finger family protein] |
0.651 |
-0.003 |
| 239 |
247649_at |
AT5G60030
|
unknown |
0.648 |
-0.001 |
| 240 |
247318_at |
AT5G63990
|
[Inositol monophosphatase family protein] |
0.648 |
0.023 |
| 241 |
250127_at |
AT5G16380
|
unknown |
0.646 |
0.071 |
| 242 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.645 |
-0.008 |
| 243 |
245078_at |
AT2G23340
|
DEAR3, DREB and EAR motif protein 3 |
0.643 |
0.122 |
| 244 |
266839_at |
AT2G25930
|
ELF3, EARLY FLOWERING 3, PYK20 |
0.642 |
-0.021 |
| 245 |
255585_at |
AT4G01550
|
ANAC069, NAC domain containing protein 69, NAC069, NAC domain containing protein 69, NTM2, NAC with Transmembrane Motif 2 |
0.642 |
0.044 |
| 246 |
265216_at |
AT1G05100
|
MAPKKK18, mitogen-activated protein kinase kinase kinase 18 |
0.641 |
0.091 |
| 247 |
254085_at |
AT4G24960
|
ATHVA22D, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE D, HVA22D, HVA22 homologue D |
0.641 |
-0.077 |
| 248 |
245427_at |
AT4G17550
|
AtG3Pp4, glycerol-3-phosphate permease 4, G3Pp4, glycerol-3-phosphate permease 4 |
0.640 |
-0.079 |
| 249 |
263222_at |
AT1G30640
|
[Protein kinase family protein] |
0.640 |
-0.009 |
| 250 |
248160_at |
AT5G54470
|
BBX29, B-box domain protein 29 |
0.639 |
0.069 |
| 251 |
262440_at |
AT1G47710
|
AtSERPIN1, SERPIN1 |
0.637 |
-0.018 |
| 252 |
245276_at |
AT4G16780
|
ATHB-2, homeobox protein 2, ATHB2, ARABIDOPSIS THALIANA HOMEOBOX PROTEIN 2, HAT4, HB-2, homeobox protein 2 |
0.634 |
0.001 |
| 253 |
245200_at |
AT1G67850
|
unknown |
0.634 |
0.095 |
| 254 |
263249_at |
AT2G31360
|
ADS2, 16:0delta9 desaturase 2, AtADS2 |
0.633 |
0.029 |
| 255 |
247774_at |
AT5G58660
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.631 |
-0.105 |
| 256 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.630 |
0.005 |
| 257 |
254193_at |
AT4G23870
|
unknown |
0.629 |
-0.001 |
| 258 |
250504_at |
AT5G09840
|
[Putative endonuclease or glycosyl hydrolase] |
0.628 |
-0.057 |
| 259 |
253614_at |
AT4G30350
|
SMXL2, SMAX1-like 2 |
0.626 |
-0.016 |
| 260 |
251346_at |
AT3G60980
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.625 |
-0.053 |
| 261 |
260345_at |
AT1G69270
|
AtRPK1, RPK1, receptor-like protein kinase 1 |
0.621 |
-0.061 |
| 262 |
265728_at |
AT2G31990
|
[Exostosin family protein] |
0.619 |
-0.004 |
| 263 |
251789_at |
AT3G55450
|
PBL1, PBS1-like 1 |
0.618 |
-0.003 |
| 264 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.613 |
0.021 |
| 265 |
263384_at |
AT2G40130
|
SMXL8, SMAX1-like 8 |
0.613 |
-0.072 |
| 266 |
253737_at |
AT4G28703
|
[RmlC-like cupins superfamily protein] |
0.612 |
0.057 |
| 267 |
258209_at |
AT3G14060
|
unknown |
0.608 |
-0.061 |
| 268 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.608 |
0.153 |
| 269 |
255070_at |
AT4G09020
|
ATISA3, ISA3, isoamylase 3 |
0.606 |
0.011 |
| 270 |
247739_at |
AT5G59240
|
[Ribosomal protein S8e family protein] |
0.605 |
-0.045 |
| 271 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.605 |
0.099 |
| 272 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.604 |
0.040 |
| 273 |
255452_at |
AT4G02880
|
unknown |
0.604 |
-0.041 |
| 274 |
250670_at |
AT5G06860
|
ATPGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1, PGIP1, polygalacturonase inhibiting protein 1 |
0.604 |
-0.109 |
| 275 |
258507_at |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
0.602 |
-0.068 |
| 276 |
266555_at |
AT2G46270
|
GBF3, G-box binding factor 3 |
0.600 |
0.044 |
| 277 |
250694_at |
AT5G06710
|
HAT14, homeobox from Arabidopsis thaliana |
0.600 |
0.039 |
| 278 |
263082_at |
AT2G27200
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.600 |
0.036 |
| 279 |
260203_at |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
0.599 |
-0.067 |
| 280 |
261004_at |
AT1G26450
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.598 |
0.083 |
| 281 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.596 |
-0.083 |
| 282 |
255236_at |
AT4G05520
|
ATEHD2, EPS15 homology domain 2, EHD2, EPS15 homology domain 2 |
0.594 |
-0.053 |
| 283 |
249411_at |
AT5G40390
|
RS5, raffinose synthase 5, SIP1, seed imbibition 1-like |
0.594 |
0.051 |
| 284 |
259426_at |
AT1G01470
|
LEA14, LATE EMBRYOGENESIS ABUNDANT 14, LSR3, LIGHT STRESS-REGULATED 3 |
0.593 |
-0.002 |
| 285 |
264818_at |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
0.590 |
-0.032 |
| 286 |
247957_at |
AT5G57050
|
ABI2, ABA INSENSITIVE 2, AtABI2 |
0.588 |
-0.017 |
| 287 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.587 |
-0.018 |
| 288 |
253828_at |
AT4G27970
|
SLAH2, SLAC1 homologue 2 |
0.586 |
0.050 |
| 289 |
245533_at |
AT4G15130
|
ATCCT2, CCT2, phosphorylcholine cytidylyltransferase2 |
0.585 |
-0.026 |
| 290 |
265478_at |
AT2G15890
|
MEE14, maternal effect embryo arrest 14 |
0.585 |
-0.043 |
| 291 |
263574_at |
AT2G16990
|
[Major facilitator superfamily protein] |
0.585 |
-0.027 |
| 292 |
245352_at |
AT4G15490
|
UGT84A3 |
0.582 |
0.113 |
| 293 |
253603_at |
AT4G30935
|
ATWRKY32, WRKY32, WRKY DNA-binding protein 32 |
0.581 |
0.004 |
| 294 |
264024_at |
AT2G21180
|
unknown |
0.580 |
0.032 |
| 295 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
0.579 |
0.161 |
| 296 |
258719_at |
AT3G09540
|
[Pectin lyase-like superfamily protein] |
0.579 |
0.121 |
| 297 |
251272_at |
AT3G61890
|
ATHB-12, homeobox 12, ATHB12, ARABIDOPSIS THALIANA HOMEOBOX 12, HB-12, homeobox 12 |
0.579 |
0.121 |
| 298 |
260338_at |
AT1G69250
|
[Nuclear transport factor 2 (NTF2) family protein with RNA binding (RRM-RBD-RNP motifs) domain] |
0.579 |
-0.051 |
| 299 |
252997_at |
AT4G38400
|
ATEXLA2, expansin-like A2, ATEXPL2, ATHEXP BETA 2.2, EXLA2, expansin-like A2, EXPL2, EXPANSIN L2 |
0.578 |
0.097 |
| 300 |
258742_at |
AT3G05800
|
AIF1, AtBS1(activation-tagged BRI1 suppressor 1)-interacting factor 1 |
0.575 |
-0.019 |
| 301 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-1.375 |
0.193 |
| 302 |
256096_at |
AT1G13650
|
unknown |
-1.256 |
-0.226 |
| 303 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-1.194 |
-0.204 |
| 304 |
253305_at |
AT4G33666
|
unknown |
-1.021 |
0.023 |
| 305 |
254574_at |
AT4G19430
|
unknown |
-1.018 |
0.028 |
| 306 |
247977_at |
AT5G56850
|
unknown |
-0.988 |
-0.039 |
| 307 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.959 |
0.259 |
| 308 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.954 |
0.024 |
| 309 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.881 |
-0.138 |
| 310 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.874 |
-0.130 |
| 311 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
-0.871 |
0.101 |
| 312 |
252433_at |
AT3G47560
|
[alpha/beta-Hydrolases superfamily protein] |
-0.867 |
-0.028 |
| 313 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
-0.863 |
-0.029 |
| 314 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.856 |
-0.052 |
| 315 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
-0.851 |
0.074 |
| 316 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-0.843 |
-0.106 |
| 317 |
253946_at |
AT4G26790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.841 |
-0.056 |
| 318 |
246021_at |
AT5G21100
|
[Plant L-ascorbate oxidase] |
-0.835 |
0.041 |
| 319 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.815 |
-0.085 |
| 320 |
253806_at |
AT4G28270
|
ATRMA2, RMA2, RING membrane-anchor 2 |
-0.800 |
0.033 |
| 321 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.798 |
0.044 |
| 322 |
247882_at |
AT5G57785
|
unknown |
-0.794 |
-0.012 |
| 323 |
264037_at |
AT2G03750
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.791 |
-0.015 |
| 324 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
-0.790 |
-0.138 |
| 325 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.790 |
0.047 |
| 326 |
256914_at |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-0.789 |
-0.053 |
| 327 |
256674_at |
AT3G52360
|
unknown |
-0.785 |
0.153 |
| 328 |
255773_at |
AT1G18590
|
ATSOT17, SULFOTRANSFERASE 17, ATST5C, ARABIDOPSIS SULFOTRANSFERASE 5C, SOT17, sulfotransferase 17 |
-0.776 |
0.123 |
| 329 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.776 |
-0.163 |
| 330 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.768 |
-0.194 |
| 331 |
248329_at |
AT5G52780
|
unknown |
-0.755 |
-0.005 |
| 332 |
266790_at |
AT2G28950
|
ATEXP6, ARABIDOPSIS THALIANA TEXPANSIN 6, ATEXPA6, expansin A6, ATHEXP ALPHA 1.8, EXPA6, expansin A6 |
-0.755 |
0.043 |
| 333 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.753 |
0.022 |
| 334 |
248683_at |
AT5G48490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.751 |
0.115 |
| 335 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.746 |
0.066 |
| 336 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
-0.741 |
0.217 |
| 337 |
262826_at |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.737 |
0.042 |
| 338 |
264507_at |
AT1G09415
|
NIMIN-3, NIM1-interacting 3 |
-0.734 |
-0.036 |
| 339 |
263674_at |
AT2G04790
|
unknown |
-0.733 |
0.015 |
| 340 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.730 |
-0.021 |
| 341 |
253886_at |
AT4G27710
|
CYP709B3, cytochrome P450, family 709, subfamily B, polypeptide 3 |
-0.726 |
0.027 |
| 342 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.725 |
0.107 |
| 343 |
253874_at |
AT4G27450
|
[Aluminium induced protein with YGL and LRDR motifs] |
-0.722 |
-0.021 |
| 344 |
266357_at |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-0.720 |
-0.023 |
| 345 |
258262_at |
AT3G15770
|
unknown |
-0.711 |
-0.122 |
| 346 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
-0.711 |
-0.055 |
| 347 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.705 |
-0.120 |
| 348 |
253412_at |
AT4G33000
|
ATCBL10, CBL10, calcineurin B-like protein 10, SCABP8, SOS3-LIKE CALCIUM BINDING PROTEIN 8 |
-0.705 |
0.003 |
| 349 |
251360_at |
AT3G61210
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.704 |
-0.035 |
| 350 |
262986_at |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-0.702 |
-0.007 |
| 351 |
248186_at |
AT5G53880
|
unknown |
-0.697 |
0.043 |
| 352 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.697 |
0.091 |
| 353 |
255579_at |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.691 |
0.025 |
| 354 |
261308_at |
AT1G48480
|
RKL1, receptor-like kinase 1 |
-0.687 |
0.114 |
| 355 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.683 |
-0.095 |
| 356 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
-0.682 |
0.032 |
| 357 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.680 |
0.033 |
| 358 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
-0.676 |
0.034 |
| 359 |
256379_at |
AT1G66840
|
PMI2, plastid movement impaired 2, WEB2, WEAK CHLOROPLAST MOVEMENT UNDER BLUE LIGHT 2 |
-0.670 |
-0.123 |
| 360 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.669 |
0.159 |
| 361 |
255774_at |
AT1G18620
|
TRM3, TON1 Recruiting Motif 3 |
-0.668 |
0.008 |
| 362 |
250478_at |
AT5G10250
|
DOT3, DEFECTIVELY ORGANIZED TRIBUTARIES 3 |
-0.666 |
-0.007 |
| 363 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.664 |
0.050 |
| 364 |
260431_at |
AT1G68190
|
BBX27, B-box domain protein 27 |
-0.663 |
0.022 |
| 365 |
246591_at |
AT5G14880
|
KUP8, potassium uptake 8 |
-0.662 |
0.026 |
| 366 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
-0.662 |
0.119 |
| 367 |
266372_at |
AT2G41310
|
ARR8, RESPONSE REGULATOR 8, ATRR3, response regulator 3, RR3, response regulator 3 |
-0.660 |
0.044 |
| 368 |
260914_at |
AT1G02640
|
ATBXL2, BETA-XYLOSIDASE 2, BXL2, beta-xylosidase 2 |
-0.660 |
-0.019 |
| 369 |
254119_at |
AT4G24780
|
[Pectin lyase-like superfamily protein] |
-0.659 |
0.004 |
| 370 |
261782_at |
AT1G76110
|
[HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain] |
-0.657 |
0.175 |
| 371 |
247040_at |
AT5G67150
|
[HXXXD-type acyl-transferase family protein] |
-0.655 |
0.105 |
| 372 |
259760_at |
AT1G77580
|
unknown |
-0.651 |
-0.021 |
| 373 |
256980_at |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.650 |
-0.048 |
| 374 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.644 |
0.159 |
| 375 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-0.641 |
0.155 |
| 376 |
253534_at |
AT4G31500
|
ATR4, ALTERED TRYPTOPHAN REGULATION 4, CYP83B1, cytochrome P450, family 83, subfamily B, polypeptide 1, RED1, RED ELONGATED 1, RNT1, RUNT 1, SUR2, SUPERROOT 2 |
-0.638 |
0.050 |
| 377 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.638 |
0.117 |
| 378 |
253048_at |
AT4G37560
|
[Acetamidase/Formamidase family protein] |
-0.634 |
-0.068 |
| 379 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
-0.632 |
-0.113 |
| 380 |
262049_at |
AT1G80180
|
unknown |
-0.628 |
-0.052 |
| 381 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
-0.627 |
0.033 |
| 382 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.626 |
0.091 |
| 383 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.618 |
0.035 |
| 384 |
249727_at |
AT5G35490
|
ATMRU1, ARABIDOPSIS MTO 1 RESPONDING UP 1, MRU1, MTO 1 RESPONDING UP 1 |
-0.617 |
0.039 |
| 385 |
257474_at |
AT1G80850
|
[DNA glycosylase superfamily protein] |
-0.616 |
0.074 |
| 386 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.615 |
-0.064 |
| 387 |
259822_at |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.614 |
0.128 |
| 388 |
246968_at |
AT5G24870
|
[RING/U-box superfamily protein] |
-0.614 |
0.048 |
| 389 |
258919_at |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-0.613 |
0.150 |
| 390 |
250008_at |
AT5G18630
|
[alpha/beta-Hydrolases superfamily protein] |
-0.611 |
0.055 |
| 391 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.611 |
0.035 |
| 392 |
247266_at |
AT5G64570
|
ATBXL4, ARABIDOPSIS THALIANA BETA-D-XYLOSIDASE 4, XYL4, beta-D-xylosidase 4 |
-0.611 |
0.001 |
| 393 |
252911_at |
AT4G39510
|
CYP96A12, cytochrome P450, family 96, subfamily A, polypeptide 12 |
-0.607 |
0.025 |
| 394 |
260969_at |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
-0.607 |
0.037 |
| 395 |
256332_at |
AT1G76890
|
AT-GT2, GT2 |
-0.606 |
0.093 |
| 396 |
263184_at |
AT1G05560
|
UGT1, UDP-GLUCOSE TRANSFERASE 1, UGT75B1, UDP-glucosyltransferase 75B1 |
-0.603 |
0.095 |
| 397 |
254145_at |
AT4G24700
|
unknown |
-0.603 |
0.037 |
| 398 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.601 |
0.075 |
| 399 |
252478_at |
AT3G46540
|
[ENTH/VHS family protein] |
-0.600 |
-0.032 |
| 400 |
254024_at |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.599 |
0.062 |
| 401 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.595 |
0.035 |
| 402 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.592 |
-0.078 |
| 403 |
262354_at |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
-0.591 |
0.052 |
| 404 |
267034_at |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-0.591 |
-0.010 |
| 405 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.591 |
0.078 |
| 406 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.590 |
0.130 |
| 407 |
255294_at |
AT4G04750
|
[Major facilitator superfamily protein] |
-0.589 |
0.059 |
| 408 |
252092_at |
AT3G51420
|
ATSSL4, STRICTOSIDINE SYNTHASE-LIKE 4, SSL4, strictosidine synthase-like 4 |
-0.589 |
-0.003 |
| 409 |
260380_at |
AT1G73870
|
BBX16, B-box domain protein 16, COL7, CONSTANS-LIKE 7 |
-0.587 |
0.078 |
| 410 |
251746_at |
AT3G56060
|
[Glucose-methanol-choline (GMC) oxidoreductase family protein] |
-0.587 |
0.100 |
| 411 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
-0.584 |
0.092 |
| 412 |
262286_at |
AT1G68585
|
unknown |
-0.583 |
0.268 |
| 413 |
258156_at |
AT3G18050
|
unknown |
-0.583 |
0.012 |
| 414 |
266591_at |
AT2G46225
|
ABIL1, ABI-1-like 1 |
-0.580 |
-0.053 |
| 415 |
253302_at |
AT4G33660
|
unknown |
-0.579 |
0.023 |
| 416 |
261500_at |
AT1G28400
|
unknown |
-0.574 |
0.004 |
| 417 |
249542_at |
AT5G38140
|
NF-YC12, nuclear factor Y, subunit C12 |
-0.572 |
-0.014 |
| 418 |
253814_at |
AT4G28290
|
unknown |
-0.570 |
0.042 |
| 419 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
-0.570 |
0.036 |
| 420 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
-0.569 |
0.111 |
| 421 |
257801_at |
AT3G18750
|
ATWNK6, ARABIDOPSIS THALIANA WITH NO K 6, WNK6, with no lysine (K) kinase 6, ZIK5 |
-0.566 |
-0.028 |
| 422 |
257510_at |
AT1G55360
|
unknown |
-0.566 |
0.009 |
| 423 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
-0.565 |
0.112 |
| 424 |
259545_at |
AT1G20560
|
AAE1, acyl activating enzyme 1 |
-0.562 |
0.057 |
| 425 |
256603_at |
AT3G28270
|
unknown |
-0.562 |
0.363 |
| 426 |
267359_at |
AT2G40020
|
[Nucleolar histone methyltransferase-related protein] |
-0.561 |
0.022 |
| 427 |
256908_at |
AT3G24040
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
-0.557 |
0.031 |
| 428 |
246576_at |
AT1G31650
|
ATROPGEF14, ROPGEF14, ROP (rho of plants) guanine nucleotide exchange factor 14 |
-0.553 |
-0.003 |
| 429 |
250017_at |
AT5G18140
|
DJC69, DNA J protein A69 |
-0.552 |
0.033 |
| 430 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.552 |
-0.047 |
| 431 |
257966_at |
AT3G19800
|
unknown |
-0.550 |
-0.019 |
| 432 |
252972_at |
AT4G38840
|
SAUR14, SMALL AUXIN UPREGULATED RNA 14 |
-0.548 |
0.006 |
| 433 |
250696_at |
AT5G06790
|
unknown |
-0.547 |
0.126 |
| 434 |
247072_at |
AT5G66490
|
unknown |
-0.545 |
0.021 |
| 435 |
263106_at |
AT2G05160
|
[CCCH-type zinc fingerfamily protein with RNA-binding domain] |
-0.545 |
0.032 |
| 436 |
260167_at |
AT1G71970
|
unknown |
-0.545 |
0.011 |
| 437 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.542 |
-0.027 |
| 438 |
246231_at |
AT4G37080
|
unknown |
-0.541 |
0.139 |
| 439 |
254133_at |
AT4G24810
|
[Protein kinase superfamily protein] |
-0.540 |
0.032 |
| 440 |
264697_at |
AT1G70210
|
ATCYCD1;1, CYCD1;1, CYCLIN D1;1 |
-0.538 |
0.062 |
| 441 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.535 |
-0.085 |
| 442 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
-0.535 |
0.162 |
| 443 |
245130_at |
AT2G45340
|
[Leucine-rich repeat protein kinase family protein] |
-0.532 |
0.038 |
| 444 |
265573_at |
AT2G28200
|
[C2H2-type zinc finger family protein] |
-0.531 |
0.017 |
| 445 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.531 |
0.007 |
| 446 |
260109_at |
AT1G63260
|
TET10, tetraspanin10 |
-0.530 |
0.011 |
| 447 |
263135_at |
AT1G78550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.530 |
-0.041 |
| 448 |
250255_at |
AT5G13730
|
SIG4, sigma factor 4, SIGD |
-0.528 |
-0.002 |
| 449 |
251856_at |
AT3G54720
|
AMP1, ALTERED MERISTEM PROGRAM 1, AtAMP1, COP2, CONSTITUTIVE MORPHOGENESIS 2, HPT, HAUPTLING, MFO1, Multifolia, PT, PRIMORDIA TIMING |
-0.528 |
0.096 |
| 450 |
262730_at |
AT1G16390
|
ATOCT3, organic cation/carnitine transporter 3, OCT3, organic cation/carnitine transporter 3 |
-0.528 |
0.000 |
| 451 |
266589_at |
AT2G46250
|
[myosin heavy chain-related] |
-0.527 |
-0.055 |
| 452 |
260317_at |
AT1G63800
|
UBC5, ubiquitin-conjugating enzyme 5 |
-0.526 |
-0.015 |
| 453 |
264001_at |
AT2G22420
|
[Peroxidase superfamily protein] |
-0.526 |
0.012 |
| 454 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.524 |
-0.001 |
| 455 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.523 |
-0.057 |
| 456 |
255857_at |
AT1G67080
|
ABA4, abscisic acid (ABA)-deficient 4 |
-0.522 |
0.011 |
| 457 |
254333_at |
AT4G22753
|
ATSMO1-3, SMO1-3, sterol 4-alpha methyl oxidase 1-3 |
-0.521 |
0.059 |
| 458 |
254794_at |
AT4G12970
|
EPFL9, STOMAGEN, STOMAGEN |
-0.519 |
0.073 |
| 459 |
246275_at |
AT4G36540
|
BEE2, BR enhanced expression 2 |
-0.519 |
-0.069 |
| 460 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.519 |
-0.062 |
| 461 |
262780_at |
AT1G13090
|
CYP71B28, cytochrome P450, family 71, subfamily B, polypeptide 28 |
-0.518 |
0.124 |
| 462 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.517 |
0.050 |
| 463 |
246997_at |
AT5G67390
|
unknown |
-0.516 |
-0.063 |
| 464 |
262811_at |
AT1G11700
|
unknown |
-0.515 |
0.176 |
| 465 |
261139_at |
AT1G19700
|
BEL10, BEL1-like homeodomain 10, BLH10, BEL1-LIKE HOMEODOMAIN 10 |
-0.515 |
0.075 |
| 466 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
-0.514 |
0.161 |
| 467 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.513 |
0.010 |
| 468 |
246200_at |
AT4G37240
|
unknown |
-0.513 |
0.022 |
| 469 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
-0.513 |
0.038 |
| 470 |
254746_at |
AT4G12980
|
[Auxin-responsive family protein] |
-0.512 |
0.084 |
| 471 |
266300_at |
AT2G01420
|
ATPIN4, ARABIDOPSIS PIN-FORMED 4, PIN4, PIN-FORMED 4 |
-0.511 |
-0.062 |
| 472 |
264229_at |
AT1G67480
|
[Galactose oxidase/kelch repeat superfamily protein] |
-0.511 |
0.004 |
| 473 |
254561_at |
AT4G19160
|
unknown |
-0.510 |
0.080 |
| 474 |
252143_at |
AT3G51150
|
[ATP binding microtubule motor family protein] |
-0.510 |
-0.053 |
| 475 |
253397_at |
AT4G32710
|
PERK14, proline-rich extensin-like receptor kinase 14 |
-0.508 |
0.124 |
| 476 |
245592_at |
AT4G14540
|
NF-YB3, nuclear factor Y, subunit B3 |
-0.508 |
0.008 |
| 477 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.507 |
-0.456 |
| 478 |
248046_at |
AT5G56040
|
SKM2, STERILITY-REGULATING KINASE MEMBER 2 |
-0.507 |
-0.042 |
| 479 |
257299_at |
AT3G28050
|
UMAMIT41, Usually multiple acids move in and out Transporters 41 |
-0.507 |
0.055 |
| 480 |
262577_at |
AT1G15290
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.507 |
-0.001 |
| 481 |
260769_at |
AT1G49010
|
[Duplicated homeodomain-like superfamily protein] |
-0.505 |
0.015 |
| 482 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
-0.505 |
-0.002 |
| 483 |
250968_at |
AT5G02890
|
[HXXXD-type acyl-transferase family protein] |
-0.505 |
-0.021 |
| 484 |
266265_at |
AT2G29340
|
[NAD-dependent epimerase/dehydratase family protein] |
-0.503 |
0.004 |
| 485 |
248868_at |
AT5G46780
|
[VQ motif-containing protein] |
-0.503 |
0.007 |
| 486 |
253548_at |
AT4G30993
|
[Calcineurin-like metallo-phosphoesterase superfamily protein] |
-0.503 |
-0.017 |
| 487 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.501 |
-0.256 |
| 488 |
247819_at |
AT5G58350
|
WNK4, with no lysine (K) kinase 4, ZIK2 |
-0.501 |
-0.009 |
| 489 |
252178_at |
AT3G50750
|
BEH1, BES1/BZR1 homolog 1 |
-0.500 |
0.046 |
| 490 |
245426_at |
AT4G17540
|
unknown |
-0.500 |
-0.023 |
| 491 |
267265_at |
AT2G22980
|
SCPL13, serine carboxypeptidase-like 13 |
-0.499 |
0.043 |
| 492 |
254226_at |
AT4G23690
|
AtDIR6, Arabidopsis thaliana dirigent protein 6, DIR6, dirigent protein 6 |
-0.499 |
0.048 |
| 493 |
267544_at |
AT2G32720
|
ATCB5-B, ARABIDOPSIS CYTOCHROME B5 ISOFORM B, B5 #4, CB5-B, cytochrome B5 isoform B, CYTB5-D |
-0.498 |
-0.015 |
| 494 |
255302_at |
AT4G04830
|
ATMSRB5, methionine sulfoxide reductase B5, MSRB5, methionine sulfoxide reductase B5 |
-0.497 |
-0.029 |
| 495 |
266391_at |
AT2G41290
|
SSL2, strictosidine synthase-like 2 |
-0.495 |
0.071 |
| 496 |
258782_at |
AT3G11750
|
FOLB1 |
-0.495 |
-0.098 |
| 497 |
264521_at |
AT1G10020
|
unknown |
-0.492 |
-0.010 |
| 498 |
255583_at |
AT4G01510
|
ARV2 |
-0.491 |
0.004 |
| 499 |
260395_at |
AT1G69780
|
ATHB13 |
-0.491 |
-0.147 |
| 500 |
248291_at |
AT5G53020
|
[Ribonuclease P protein subunit P38-related] |
-0.490 |
0.061 |
| 501 |
261016_at |
AT1G26560
|
BGLU40, beta glucosidase 40 |
-0.489 |
0.008 |
| 502 |
257314_at |
AT3G26590
|
[MATE efflux family protein] |
-0.489 |
-0.060 |
| 503 |
267344_at |
AT2G44230
|
unknown |
-0.488 |
0.027 |
| 504 |
248153_at |
AT5G54250
|
ATCNGC4, cyclic nucleotide-gated cation channel 4, CNGC4, cyclic nucleotide-gated cation channel 4, DND2, DEFENSE, NO DEATH 2, HLM1 |
-0.487 |
0.016 |
| 505 |
259954_at |
AT1G75130
|
CYP721A1, cytochrome P450, family 721, subfamily A, polypeptide 1 |
-0.487 |
-0.022 |
| 506 |
261803_at |
AT1G30500
|
NF-YA7, nuclear factor Y, subunit A7 |
-0.486 |
0.032 |
| 507 |
245984_at |
AT5G13090
|
unknown |
-0.485 |
-0.009 |
| 508 |
264826_at |
AT1G03410
|
2A6 |
-0.485 |
0.056 |
| 509 |
264770_at |
AT1G23030
|
[ARM repeat superfamily protein] |
-0.485 |
0.027 |
| 510 |
254803_at |
AT4G13100
|
[RING/U-box superfamily protein] |
-0.484 |
0.074 |
| 511 |
247946_at |
AT5G57180
|
CIA2, chloroplast import apparatus 2 |
-0.483 |
-0.078 |
| 512 |
260592_at |
AT1G55850
|
ATCSLE1, CSLE1, cellulose synthase like E1 |
-0.482 |
-0.014 |
| 513 |
260983_at |
AT1G53560
|
[Ribosomal protein L18ae family] |
-0.482 |
0.067 |
| 514 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
-0.480 |
0.060 |
| 515 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
-0.479 |
0.104 |
| 516 |
261485_at |
AT1G14360
|
ATUTR3, UTR3, UDP-galactose transporter 3 |
-0.477 |
-0.011 |
| 517 |
263647_at |
AT2G04690
|
Pyridoxamine 5'-phosphate oxidase family protein |
-0.477 |
0.012 |
| 518 |
254705_at |
AT4G17870
|
PYR1, PYRABACTIN RESISTANCE 1, RCAR11, regulatory component of ABA receptor 11 |
-0.477 |
0.090 |
| 519 |
259540_at |
AT1G20640
|
NLP4, NIN-like protein 4 |
-0.476 |
0.045 |
| 520 |
257203_at |
AT3G23730
|
XTH16, xyloglucan endotransglucosylase/hydrolase 16 |
-0.476 |
0.029 |
| 521 |
254851_at |
AT4G12010
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.475 |
0.053 |
| 522 |
263866_at |
AT2G36950
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.475 |
0.008 |
| 523 |
246487_at |
AT5G16030
|
unknown |
-0.473 |
0.020 |
| 524 |
251391_at |
AT3G60910
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.471 |
-0.004 |
| 525 |
265443_at |
AT2G20750
|
ATEXPB1, expansin B1, ATHEXP BETA 1.5, EXPB1, expansin B1 |
-0.471 |
0.338 |
| 526 |
246759_at |
AT5G27950
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.470 |
-0.113 |
| 527 |
247187_at |
AT5G65490
|
unknown |
-0.469 |
-0.048 |
| 528 |
256599_at |
AT3G14760
|
unknown |
-0.468 |
0.006 |
| 529 |
260388_at |
AT1G74070
|
[Cyclophilin-like peptidyl-prolyl cis-trans isomerase family protein] |
-0.467 |
0.047 |
| 530 |
256892_at |
AT3G19000
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.466 |
-0.006 |
| 531 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
-0.465 |
-0.002 |
| 532 |
256721_at |
AT2G34150
|
ATRANGAP2, ATSCAR1, SCAR1, WAVE1, WISKOTT-ALDRICH SYNDROME PROTEIN FAMILY VERPROLIN HOMOLOGOUS PROTEIN 1 |
-0.465 |
0.042 |
| 533 |
265342_at |
AT2G18300
|
HBI1, HOMOLOG OF BEE2 INTERACTING WITH IBH 1 |
-0.464 |
-0.017 |
| 534 |
256427_at |
AT3G11090
|
LBD21, LOB domain-containing protein 21 |
-0.462 |
-0.023 |
| 535 |
249190_at |
AT5G42750
|
BKI1, BRI1 kinase inhibitor 1 |
-0.461 |
-0.018 |
| 536 |
267347_at |
AT2G39950
|
unknown |
-0.461 |
0.007 |
| 537 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-0.459 |
0.148 |
| 538 |
251142_at |
AT5G01015
|
unknown |
-0.458 |
0.134 |
| 539 |
257999_at |
AT3G27540
|
[beta-1,4-N-acetylglucosaminyltransferase family protein] |
-0.458 |
0.082 |
| 540 |
263777_at |
AT2G46450
|
ATCNGC12, cyclic nucleotide-gated channel 12, CNGC12, cyclic nucleotide-gated channel 12 |
-0.458 |
0.004 |
| 541 |
250110_at |
AT5G15350
|
AtENODL17, ENODL17, early nodulin-like protein 17 |
-0.457 |
0.017 |
| 542 |
257547_at |
AT3G13000
|
unknown |
-0.456 |
-0.045 |
| 543 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
-0.456 |
0.233 |
| 544 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
-0.456 |
0.006 |
| 545 |
252652_at |
AT3G44720
|
ADT4, arogenate dehydratase 4 |
-0.455 |
0.067 |
| 546 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
-0.455 |
0.132 |
| 547 |
261712_at |
AT1G32780
|
[GroES-like zinc-binding dehydrogenase family protein] |
-0.455 |
0.061 |
| 548 |
256880_at |
AT3G26450
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.454 |
0.041 |
| 549 |
259434_at |
AT1G01490
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.454 |
-0.031 |
| 550 |
260266_at |
AT1G68520
|
BBX14, B-box domain protein 14 |
-0.454 |
0.046 |
| 551 |
254532_at |
AT4G19660
|
ATNPR4, NPR4, NPR1-like protein 4 |
-0.452 |
-0.024 |
| 552 |
259853_at |
AT1G72300
|
PSY1R, PSY1 receptor |
-0.452 |
-0.029 |
| 553 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
-0.451 |
0.011 |
| 554 |
249694_at |
AT5G35790
|
G6PD1, glucose-6-phosphate dehydrogenase 1 |
-0.451 |
0.051 |
| 555 |
262411_at |
AT1G34640
|
[peptidases] |
-0.450 |
0.129 |
| 556 |
254938_at |
AT4G10770
|
ATOPT7, ARABIDOPSIS THALIANA OLIGOPEPTIDE TRANSPORTER 7, OPT7, oligopeptide transporter 7 |
-0.450 |
0.055 |
| 557 |
262705_at |
AT1G16260
|
[Wall-associated kinase family protein] |
-0.449 |
-0.057 |
| 558 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.449 |
0.120 |
| 559 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
-0.448 |
0.063 |
| 560 |
254787_at |
AT4G12690
|
unknown |
-0.448 |
-0.103 |
| 561 |
265990_at |
AT2G24280
|
[alpha/beta-Hydrolases superfamily protein] |
-0.447 |
0.075 |
| 562 |
262563_at |
AT1G34210
|
ATSERK2, SERK2, somatic embryogenesis receptor-like kinase 2 |
-0.446 |
0.004 |
| 563 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.446 |
0.034 |
| 564 |
257209_at |
AT3G14920
|
[Peptide-N4-(N-acetyl-beta-glucosaminyl)asparagine amidase A protein] |
-0.446 |
-0.045 |
| 565 |
259775_at |
AT1G29530
|
unknown |
-0.445 |
0.014 |
| 566 |
254982_at |
AT4G10470
|
unknown |
-0.445 |
-0.157 |
| 567 |
260363_at |
AT1G70550
|
unknown |
-0.445 |
-0.023 |
| 568 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
-0.445 |
0.011 |
| 569 |
255310_at |
AT4G04955
|
ALN, allantoinase, ATALN, allantoinase |
-0.445 |
0.023 |
| 570 |
263946_at |
AT2G36000
|
EMB3114, EMBRYO DEFECTIVE 3114 |
-0.444 |
-0.002 |
| 571 |
264240_at |
AT1G54820
|
[Protein kinase superfamily protein] |
-0.443 |
0.100 |
| 572 |
256400_at |
AT3G06140
|
LUL4, LOG2-LIKE UBIQUITIN LIGASE4 |
-0.443 |
0.061 |
| 573 |
267380_at |
AT2G26170
|
CYP711A1, cytochrome P450, family 711, subfamily A, polypeptide 1, MAX1, MORE AXILLARY BRANCHES 1 |
-0.443 |
0.028 |
| 574 |
257789_at |
AT3G27020
|
AtYSL6, YSL6, YELLOW STRIPE like 6 |
-0.443 |
0.001 |
| 575 |
246901_at |
AT5G25630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.443 |
-0.000 |
| 576 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
-0.442 |
-0.003 |
| 577 |
261060_at |
AT1G17340
|
[Phosphoinositide phosphatase family protein] |
-0.442 |
-0.038 |
| 578 |
265726_at |
AT2G32010
|
CVL1, CVP2 like 1 |
-0.440 |
-0.045 |
| 579 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
-0.439 |
0.028 |
| 580 |
258003_at |
AT3G29030
|
ATEXP5, ARABIDOPSIS THALIANA EXPANSIN 5, ATEXPA5, ARABIDOPSIS THALIANA EXPANSIN A5, ATHEXP ALPHA 1.4, EXP5, EXPANSIN 5, EXPA5, expansin A5 |
-0.439 |
0.096 |
| 581 |
263591_at |
AT2G01910
|
ATMAP65-6, MAP65-6 |
-0.439 |
0.061 |
| 582 |
266291_at |
AT2G29320
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.439 |
0.012 |
| 583 |
253966_at |
AT4G26520
|
AtFBA7, FBA7, fructose-bisphosphate aldolase 7 |
-0.439 |
0.043 |
| 584 |
254649_at |
AT4G18570
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.438 |
0.048 |
| 585 |
253856_at |
AT4G28100
|
unknown |
-0.438 |
0.090 |
| 586 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
-0.437 |
0.068 |
| 587 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.436 |
0.007 |
| 588 |
253580_at |
AT4G30400
|
[RING/U-box superfamily protein] |
-0.436 |
0.061 |
| 589 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.435 |
0.102 |
| 590 |
265985_at |
AT2G24220
|
ATPUP5, purine permease 5, PUP5, purine permease 5 |
-0.435 |
0.009 |
| 591 |
262039_at |
AT1G80050
|
APT2, adenine phosphoribosyl transferase 2, ATAPT2, PHT1.1 |
-0.434 |
0.109 |
| 592 |
261031_at |
AT1G17360
|
unknown |
-0.434 |
0.011 |
| 593 |
249693_at |
AT5G35750
|
AHK2, histidine kinase 2, HK2, histidine kinase 2 |
-0.431 |
-0.074 |
| 594 |
253270_at |
AT4G34160
|
CYCD3, CYCD3;1, CYCLIN D3;1 |
-0.431 |
0.025 |
| 595 |
262871_at |
AT1G65010
|
[INVOLVED IN: flower development] |
-0.431 |
0.021 |
| 596 |
246462_at |
AT5G16940
|
[carbon-sulfur lyases] |
-0.429 |
-0.089 |
| 597 |
260922_at |
AT1G21560
|
unknown |
-0.428 |
-0.008 |
| 598 |
264920_at |
AT1G60550
|
DHNS, ECHID, enoyl-CoA hydratase/isomerase D |
-0.427 |
0.005 |
| 599 |
246681_at |
AT5G33280
|
[Voltage-gated chloride channel family protein] |
-0.426 |
0.034 |
| 600 |
254810_at |
AT4G12390
|
PME1, pectin methylesterase inhibitor 1 |
-0.426 |
0.001 |