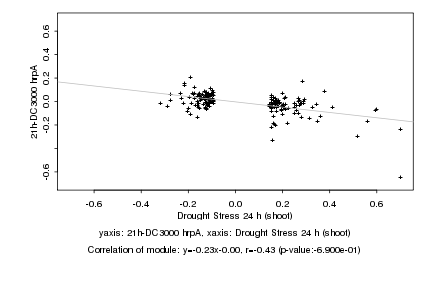
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Drought Stress 24 h (shoot) |
Log2 signal ratio
21h-DC3000 hrpA |
| 1 |
266142_at |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
0.699 |
-0.231 |
| 2 |
251770_at |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
0.698 |
-0.641 |
| 3 |
261135_at |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
0.598 |
-0.062 |
| 4 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.594 |
-0.074 |
| 5 |
254232_at |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
0.558 |
-0.167 |
| 6 |
250793_at |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.518 |
-0.290 |
| 7 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.409 |
-0.043 |
| 8 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
0.378 |
0.087 |
| 9 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
0.359 |
-0.126 |
| 10 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.345 |
-0.169 |
| 11 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.340 |
-0.020 |
| 12 |
252387_at |
AT3G47800
|
[Galactose mutarotase-like superfamily protein] |
0.325 |
-0.046 |
| 13 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.313 |
-0.144 |
| 14 |
252882_at |
AT4G39675
|
unknown |
0.293 |
0.024 |
| 15 |
264625_at |
AT1G09020
|
ATSNF4, SNF4, homolog of yeast sucrose nonfermenting 4 |
0.287 |
0.003 |
| 16 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
0.287 |
-0.012 |
| 17 |
252265_at |
AT3G49620
|
DIN11, DARK INDUCIBLE 11 |
0.284 |
0.175 |
| 18 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.279 |
-0.131 |
| 19 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.278 |
-0.012 |
| 20 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.274 |
-0.016 |
| 21 |
258663_at |
AT3G08670
|
unknown |
0.270 |
0.012 |
| 22 |
247794_at |
AT5G58670
|
ATPLC, ARABIDOPSIS THALIANA PHOSPHOLIPASE C, ATPLC1, phospholipase C1, PLC1, phospholipase C 1, PLC1, phospholipase C1 |
0.269 |
-0.030 |
| 23 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.267 |
-0.098 |
| 24 |
252183_at |
AT3G50740
|
UGT72E1, UDP-glucosyl transferase 72E1 |
0.267 |
0.032 |
| 25 |
261221_at |
AT1G19960
|
unknown |
0.266 |
0.002 |
| 26 |
252515_at |
AT3G46230
|
ATHSP17.4, ARABIDOPSIS THALIANA HEAT SHOCK PROTEIN 17.4, HSP17.4, heat shock protein 17.4 |
0.257 |
-0.029 |
| 27 |
255362_at |
AT4G04000
|
[non-LTR retrotransposon family (LINE), has a 2.6e-39 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
0.255 |
-0.075 |
| 28 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
0.250 |
-0.009 |
| 29 |
252076_at |
AT3G51660
|
[Tautomerase/MIF superfamily protein] |
0.248 |
-0.094 |
| 30 |
252618_at |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
0.247 |
-0.048 |
| 31 |
251842_at |
AT3G54580
|
[Proline-rich extensin-like family protein] |
0.221 |
-0.056 |
| 32 |
245885_at |
AT5G09440
|
EXL4, EXORDIUM like 4 |
0.218 |
-0.181 |
| 33 |
266098_at |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.211 |
-0.019 |
| 34 |
256098_at |
AT1G13700
|
PGL1, 6-phosphogluconolactonase 1 |
0.211 |
-0.060 |
| 35 |
265844_at |
AT2G35620
|
FEI2, FEI 2 |
0.210 |
0.042 |
| 36 |
260943_at |
AT1G45145
|
ATH5, THIOREDOXIN H-TYPE 5, ATTRX5, thioredoxin H-type 5, LIV1, LOCUS OF INSENSITIVITY TO VICTORIN 1, TRX-h5, THIOREDOXIN H-TYPE 5, TRX5, thioredoxin H-type 5 |
0.209 |
-0.068 |
| 37 |
267128_at |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
0.208 |
-0.044 |
| 38 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.205 |
0.029 |
| 39 |
251704_at |
AT3G56360
|
unknown |
0.203 |
-0.049 |
| 40 |
266993_at |
AT2G39210
|
[Major facilitator superfamily protein] |
0.200 |
-0.018 |
| 41 |
267209_at |
AT2G30930
|
unknown |
0.198 |
-0.028 |
| 42 |
255786_at |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
0.198 |
-0.103 |
| 43 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
0.197 |
0.075 |
| 44 |
258308_at |
AT3G30570
|
[non-LTR retrotransposon family (LINE), has a 7.9e-38 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
0.195 |
-0.068 |
| 45 |
251942_at |
AT3G53480
|
ABCG37, ATP-binding cassette G37, ATPDR9, PLEIOTROPIC DRUG RESISTANCE 9, PDR9, pleiotropic drug resistance 9, PIS1, polar auxin transport inhibitor sensitive 1 |
0.194 |
-0.069 |
| 46 |
265481_at |
AT2G15960
|
unknown |
0.193 |
-0.039 |
| 47 |
253722_at |
AT4G29190
|
AtC3H49, AtOZF2, Arabidopsis thaliana oxidation-related zinc finger 2, AtTZF3, OZF2, oxidation-related zinc finger 2, TZF3, tandem zinc finger 3 |
0.186 |
-0.008 |
| 48 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
0.185 |
-0.005 |
| 49 |
249988_at |
AT5G18310
|
unknown |
0.185 |
-0.016 |
| 50 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.184 |
-0.005 |
| 51 |
262238_at |
AT1G48300
|
DGAT3, diacylglycerol acyltransferase 3 |
0.181 |
-0.039 |
| 52 |
257397_at |
AT2G20430
|
RIC6, ROP-interactive CRIB motif-containing protein 6 |
0.180 |
0.017 |
| 53 |
246782_at |
AT5G27320
|
ATGID1C, GID1C, GA INSENSITIVE DWARF1C |
0.178 |
0.005 |
| 54 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
0.178 |
-0.034 |
| 55 |
260211_at |
AT1G74440
|
unknown |
0.175 |
-0.035 |
| 56 |
249505_at |
AT5G38870
|
[pseudogene] |
0.174 |
-0.048 |
| 57 |
247444_at |
AT5G62630
|
HIPL2, hipl2 protein precursor |
0.174 |
-0.002 |
| 58 |
263957_at |
AT2G35880
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.171 |
0.011 |
| 59 |
253629_at |
AT4G30450
|
[glycine-rich protein] |
0.171 |
-0.077 |
| 60 |
245296_at |
AT4G16370
|
oligopeptide transporter |
0.171 |
-0.011 |
| 61 |
247619_at |
AT5G60290
|
unknown |
0.170 |
0.016 |
| 62 |
250404_at |
AT5G10960
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
0.169 |
0.002 |
| 63 |
251169_at |
AT3G63210
|
MARD1, MEDIATOR OF ABA-REGULATED DORMANCY 1 |
0.169 |
0.029 |
| 64 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
0.169 |
-0.038 |
| 65 |
249790_at |
AT5G24290
|
MEB2, MEMBRANE OF ER BODY 2 |
0.168 |
-0.197 |
| 66 |
256665_at |
AT3G20700
|
[F-box associated ubiquitination effector family protein] |
0.167 |
-0.019 |
| 67 |
253666_at |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
0.167 |
-0.035 |
| 68 |
259553_x_at |
AT1G21310
|
ATEXT3, extensin 3, EXT3, extensin 3, RSH, ROOT-SHOOT-HYPOCOTYL DEFECTIVE |
0.164 |
-0.050 |
| 69 |
263032_at |
AT1G23850
|
unknown |
0.164 |
-0.189 |
| 70 |
262899_at |
AT1G59870
|
ABCG36, ATP-binding cassette G36, ATABCG36, Arabidopsis thaliana ATP-binding cassette G36, ATPDR8, ARABIDOPSIS PLEIOTROPIC DRUG RESISTANCE 8, PDR8, PLEIOTROPIC DRUG RESISTANCE 8, PEN3, PENETRATION 3 |
0.162 |
-0.011 |
| 71 |
252885_at |
AT4G39260
|
ATGRP8, GLYCINE-RICH PROTEIN 8, CCR1, cold, circadian rhythm, and RNA binding 1, GR-RBP8, glycine-rich RNA-binding protein 8, GRP8 |
0.161 |
0.038 |
| 72 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.160 |
-0.127 |
| 73 |
247030_at |
AT5G67210
|
IRX15-L, IRX15-LIKE |
0.159 |
-0.184 |
| 74 |
250032_at |
AT5G18170
|
GDH1, glutamate dehydrogenase 1 |
0.159 |
0.009 |
| 75 |
263132_at |
AT1G78560
|
[Sodium Bile acid symporter family] |
0.156 |
-0.025 |
| 76 |
267147_at |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.154 |
-0.328 |
| 77 |
248382_at |
AT5G51890
|
[Peroxidase superfamily protein] |
0.153 |
-0.052 |
| 78 |
248244_at |
AT5G53635
|
[F-box/RNI-like/FBD-like domains-containing protein] |
0.153 |
0.020 |
| 79 |
255702_at |
AT4G00230
|
XSP1, xylem serine peptidase 1 |
0.153 |
-0.034 |
| 80 |
253185_at |
AT4G35240
|
unknown |
0.152 |
0.057 |
| 81 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.152 |
-0.219 |
| 82 |
251225_at |
AT3G62660
|
GATL7, galacturonosyltransferase-like 7 |
0.151 |
-0.003 |
| 83 |
266500_at |
AT2G06925
|
ATSPLA2-ALPHA, PHOSPHOLIPASE A2-ALPHA, PLA2-ALPHA |
0.151 |
0.041 |
| 84 |
246834_at |
AT5G26630
|
[MADS-box transcription factor family protein] |
0.150 |
-0.078 |
| 85 |
249346_at |
AT5G40780
|
LHT1, lysine histidine transporter 1 |
0.150 |
-0.004 |
| 86 |
267514_at |
AT2G45630
|
[D-isomer specific 2-hydroxyacid dehydrogenase family protein] |
0.149 |
-0.045 |
| 87 |
248945_at |
AT5G45510
|
[Leucine-rich repeat (LRR) family protein] |
0.149 |
-0.005 |
| 88 |
258063_at |
AT3G14620
|
CYP72A8, cytochrome P450, family 72, subfamily A, polypeptide 8 |
0.148 |
-0.029 |
| 89 |
253866_at |
AT4G27480
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.146 |
-0.033 |
| 90 |
265597_at |
AT2G20142
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.143 |
-0.031 |
| 91 |
262412_at |
AT1G34760
|
GF14 OMICRON, GRF11, general regulatory factor 11, RHS5, ROOT HAIR SPECIFIC 5 |
0.143 |
-0.018 |
| 92 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
0.143 |
-0.035 |
| 93 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
-0.319 |
-0.014 |
| 94 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.290 |
-0.034 |
| 95 |
263840_at |
AT2G36885
|
unknown |
-0.277 |
0.068 |
| 96 |
262737_at |
AT1G28560
|
SRD2, SHOOT REDIFFERENTIATION DEFECTIVE 2 |
-0.276 |
0.012 |
| 97 |
264257_at |
AT1G09230
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.234 |
0.074 |
| 98 |
266384_at |
AT2G14660
|
unknown |
-0.229 |
0.033 |
| 99 |
264638_at |
AT1G65480
|
FT, FLOWERING LOCUS T, RSB8, REDUCED STEM BRANCHING 8 |
-0.221 |
-0.014 |
| 100 |
250685_at |
AT5G06670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-0.219 |
0.138 |
| 101 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
-0.218 |
0.154 |
| 102 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
-0.204 |
-0.080 |
| 103 |
250566_at |
AT5G08070
|
TCP17, TCP domain protein 17 |
-0.203 |
-0.054 |
| 104 |
248049_at |
AT5G56090
|
COX15, cytochrome c oxidase 15 |
-0.198 |
0.036 |
| 105 |
253496_at |
AT4G31870
|
ATGPX7, GLUTATHIONE PEROXIDASE 7, GPX7, glutathione peroxidase 7 |
-0.194 |
-0.106 |
| 106 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-0.192 |
0.207 |
| 107 |
247293_at |
AT5G64510
|
TIN1, tunicamycin induced 1 |
-0.187 |
-0.015 |
| 108 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
-0.184 |
0.069 |
| 109 |
256619_at |
AT3G24460
|
[Serinc-domain containing serine and sphingolipid biosynthesis protein] |
-0.181 |
0.062 |
| 110 |
266668_at |
AT2G29760
|
OTP81, ORGANELLE TRANSCRIPT PROCESSING 81 |
-0.178 |
0.126 |
| 111 |
263593_at |
AT2G01860
|
EMB975, EMBRYO DEFECTIVE 975 |
-0.178 |
0.026 |
| 112 |
258474_at |
AT3G02650
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.176 |
0.076 |
| 113 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.173 |
-0.029 |
| 114 |
246765_at |
AT5G27330
|
[Prefoldin chaperone subunit family protein] |
-0.165 |
0.051 |
| 115 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.165 |
-0.136 |
| 116 |
265665_at |
AT2G27420
|
[Cysteine proteinases superfamily protein] |
-0.163 |
0.003 |
| 117 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.163 |
-0.027 |
| 118 |
261750_at |
AT1G76120
|
[Pseudouridine synthase family protein] |
-0.161 |
0.064 |
| 119 |
245364_at |
AT4G15790
|
unknown |
-0.160 |
-0.009 |
| 120 |
246540_at |
AT5G15600
|
SP1L4, SPIRAL1-like4 |
-0.158 |
-0.048 |
| 121 |
262491_at |
AT1G21650
|
SECA2 |
-0.156 |
0.076 |
| 122 |
256303_at |
AT1G69550
|
[disease resistance protein (TIR-NBS-LRR class)] |
-0.154 |
0.054 |
| 123 |
254572_at |
AT4G19380
|
[Long-chain fatty alcohol dehydrogenase family protein] |
-0.153 |
0.049 |
| 124 |
247605_at |
AT5G60970
|
TCP5, TEOSINTE BRANCHED 1, cycloidea and PCF transcription factor 5 |
-0.153 |
-0.054 |
| 125 |
265682_at |
AT2G24390
|
[AIG2-like (avirulence induced gene) family protein] |
-0.149 |
0.029 |
| 126 |
255647_at |
AT4G00900
|
ATECA2, ARABIDOPSIS THALIANA ER-TYPE CA2+-ATPASE 2, ECA2, ER-type Ca2+-ATPase 2 |
-0.149 |
0.017 |
| 127 |
257909_at |
AT3G25480
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
-0.146 |
0.042 |
| 128 |
256926_at |
AT3G22540
|
unknown |
-0.145 |
0.020 |
| 129 |
251001_at |
AT5G02670
|
unknown |
-0.144 |
0.062 |
| 130 |
255142_at |
AT4G08390
|
SAPX, stromal ascorbate peroxidase |
-0.139 |
0.035 |
| 131 |
261914_at |
AT1G65870
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
-0.136 |
-0.025 |
| 132 |
262236_at |
AT1G48330
|
unknown |
-0.135 |
0.088 |
| 133 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.134 |
0.023 |
| 134 |
246899_at |
AT5G25590
|
[INVOLVED IN: N-terminal protein myristoylation] |
-0.132 |
0.031 |
| 135 |
245055_at |
AT2G26470
|
unknown |
-0.131 |
0.079 |
| 136 |
248899_at |
AT5G46390
|
[Peptidase S41 family protein] |
-0.131 |
-0.051 |
| 137 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.130 |
-0.045 |
| 138 |
260987_at |
AT1G53590
|
NTMC2T6.1, NTMC2TYPE6.1 |
-0.129 |
-0.005 |
| 139 |
247097_at |
AT5G66460
|
AtMAN7, MAN7, endo-beta-mannase 7 |
-0.129 |
0.018 |
| 140 |
266982_at |
AT2G39500
|
unknown |
-0.128 |
0.024 |
| 141 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
-0.128 |
0.053 |
| 142 |
250421_at |
AT5G11270
|
OCP3, overexpressor of cationic peroxidase 3 |
-0.127 |
0.083 |
| 143 |
264444_at |
AT1G27360
|
SPL11, squamosa promoter-like 11 |
-0.126 |
-0.055 |
| 144 |
263674_at |
AT2G04790
|
unknown |
-0.125 |
-0.005 |
| 145 |
257357_at |
AT2G41050
|
[PQ-loop repeat family protein / transmembrane family protein] |
-0.124 |
-0.014 |
| 146 |
258019_at |
AT3G19470
|
[F-box and associated interaction domains-containing protein] |
-0.124 |
0.051 |
| 147 |
249588_at |
AT5G37790
|
[Protein kinase superfamily protein] |
-0.124 |
0.078 |
| 148 |
253814_at |
AT4G28290
|
unknown |
-0.123 |
-0.067 |
| 149 |
263621_at |
AT2G04630
|
NRPB6B, NRPE6B |
-0.122 |
0.037 |
| 150 |
257001_at |
AT3G14080
|
LSM1B, SM-like 1B |
-0.122 |
-0.015 |
| 151 |
264201_at |
AT1G22630
|
unknown |
-0.121 |
-0.032 |
| 152 |
250715_at |
AT5G06160
|
ATO, ATROPOS |
-0.121 |
0.047 |
| 153 |
260727_at |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.119 |
0.060 |
| 154 |
245743_at |
AT1G51080
|
unknown |
-0.118 |
0.064 |
| 155 |
257765_at |
AT3G23020
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.118 |
0.076 |
| 156 |
263104_at |
AT2G05120
|
[Nucleoporin, Nup133/Nup155-like] |
-0.118 |
0.057 |
| 157 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.118 |
0.053 |
| 158 |
257352_at |
AT2G34900
|
GTE01, GTE1, GLOBAL TRANSCRIPTION FACTOR GROUP E1, IMB1, IMBIBITION-INDUCIBLE 1 |
-0.117 |
0.032 |
| 159 |
246309_at |
AT3G51790
|
AtCCME, A. thaliana homolog of E. coli CcmE, ATG1, transmembrane protein G1P-related 1, G1, transmembrane protein G1P-related 1 |
-0.117 |
-0.010 |
| 160 |
249063_at |
AT5G44110
|
ABCI21, ATP-binding cassette A21, ATNAP2, ARABIDOPSIS THALIANA NON-INTRINSIC ABC PROTEIN, ATPOP1, POP1 |
-0.117 |
-0.012 |
| 161 |
260020_at |
AT1G29990
|
PFD6, prefoldin 6 |
-0.115 |
0.044 |
| 162 |
250150_at |
AT5G14710
|
unknown |
-0.115 |
-0.012 |
| 163 |
260605_at |
AT2G43780
|
unknown |
-0.114 |
0.059 |
| 164 |
264126_at |
AT1G79280
|
AtTPR, TRANSLOCATED PROMOTER REGION, NUA, nuclear pore anchor |
-0.113 |
0.023 |
| 165 |
266333_at |
AT2G32410
|
AXL, AXR1-like, AXL1, AXR1- like 1 |
-0.112 |
-0.041 |
| 166 |
257018_at |
AT3G19630
|
[Radical SAM superfamily protein] |
-0.112 |
-0.013 |
| 167 |
258690_at |
AT3G07960
|
PIP5K6, phosphatidylinositol-4-phosphate 5-kinase 6 |
-0.112 |
0.011 |
| 168 |
264098_at |
AT1G79260
|
unknown |
-0.109 |
0.024 |
| 169 |
262245_at |
AT1G48240
|
ATNPSN12, NPSN12, novel plant snare 12 |
-0.107 |
-0.006 |
| 170 |
254982_at |
AT4G10470
|
unknown |
-0.106 |
0.055 |
| 171 |
263677_at |
AT1G04520
|
PDLP2, plasmodesmata-located protein 2 |
-0.106 |
0.115 |
| 172 |
245165_at |
AT2G33180
|
unknown |
-0.102 |
0.000 |
| 173 |
262086_at |
AT1G56050
|
[GTP-binding protein-related] |
-0.102 |
0.070 |
| 174 |
247972_at |
AT5G56740
|
HAC07, HAC7, HAG02, HAG2, histone acetyltransferase of the GNAT family 2 |
-0.102 |
0.054 |
| 175 |
246581_at |
AT1G31760
|
[SWIB/MDM2 domain superfamily protein] |
-0.100 |
0.022 |
| 176 |
264381_at |
AT2G25100
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
-0.100 |
0.026 |
| 177 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
-0.099 |
0.096 |
| 178 |
264913_at |
AT1G60770
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.098 |
0.068 |
| 179 |
257746_at |
AT3G29200
|
ATCM1, ARABIDOPSIS THALIANA CHORISMATE MUTASE 1, CM1, chorismate mutase 1 |
-0.097 |
0.014 |
| 180 |
253579_at |
AT4G30610
|
BRS1, BRI1 SUPPRESSOR 1, SCPL24, SERINE CARBOXYPEPTIDASE 24 PRECURSOR |
-0.097 |
0.056 |
| 181 |
252916_at |
AT4G38950
|
[ATP binding microtubule motor family protein] |
-0.097 |
0.078 |
| 182 |
248055_at |
AT5G55830
|
LecRK-S.7, L-type lectin receptor kinase S.7 |
-0.096 |
0.006 |
| 183 |
246120_at |
AT5G20360
|
Phox3, Phox3 |
-0.096 |
-0.009 |