|
probeID |
AGICode |
Annotation |
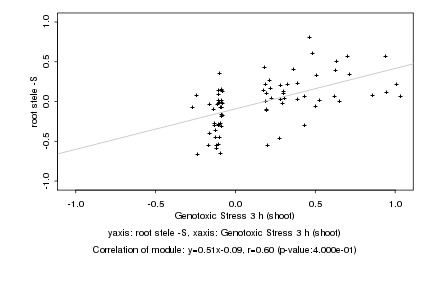
Log2 signal ratio
Genotoxic Stress 3 h (shoot) |
Log2 signal ratio
root stele -S |
| 1 |
253240_at |
AT4G34510
|
KCS17, 3-ketoacyl-CoA synthase 17 |
1.031 |
0.071 |
| 2 |
248668_at |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
1.006 |
0.216 |
| 3 |
250914_at |
AT5G03780
|
TRFL10, TRF-like 10 |
0.944 |
0.123 |
| 4 |
254443_at |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
0.937 |
0.568 |
| 5 |
255500_at |
AT4G02390
|
APP, poly(ADP-ribose) polymerase, PARP2, poly(ADP-ribose) polymerase 2, PP, poly(ADP-ribose) polymerase |
0.854 |
0.084 |
| 6 |
246132_at |
AT5G20850
|
ATRAD51, RAD51 |
0.712 |
0.349 |
| 7 |
254565_at |
AT4G19130
|
[Replication factor-A protein 1-related] |
0.698 |
0.568 |
| 8 |
249784_at |
AT5G24280
|
GMI1, GAMMA-IRRADIATION AND MITOMYCIN C INDUCED 1 |
0.650 |
0.005 |
| 9 |
249809_at |
AT5G23910
|
[ATP binding microtubule motor family protein] |
0.630 |
0.514 |
| 10 |
245137_at |
AT2G45460
|
[SMAD/FHA domain-containing protein ] |
0.621 |
0.398 |
| 11 |
255872_at |
AT2G30360
|
CIPK11, CBL-INTERACTING PROTEIN KINASE 11, PKS5, PROTEIN KINASE SOS2-LIKE 5, SIP4, SOS3-interacting protein 4, SNRK3.22, SNF1-RELATED PROTEIN KINASE 3.22 |
0.620 |
0.069 |
| 12 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
0.521 |
0.022 |
| 13 |
249326_at |
AT5G40840
|
AtRAD21.1, Sister chromatid cohesion 1 (SCC1) protein homolog 2, SYN2 |
0.504 |
0.331 |
| 14 |
262548_at |
AT1G31280
|
AGO2, argonaute 2, AtAGO2 |
0.496 |
-0.060 |
| 15 |
253051_at |
AT4G37490
|
CYC1, CYCLIN 1, CYCB1, CYCB1;1, CYCLIN B1;1 |
0.477 |
0.608 |
| 16 |
258476_at |
AT3G02400
|
[SMAD/FHA domain-containing protein ] |
0.461 |
0.806 |
| 17 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.430 |
-0.289 |
| 18 |
259224_at |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
0.427 |
0.072 |
| 19 |
258091_at |
AT3G14560
|
unknown |
0.387 |
0.036 |
| 20 |
247081_at |
AT5G66130
|
ATRAD17, RADIATION SENSITIVE 17, RAD17, RADIATION SENSITIVE 17 |
0.386 |
0.231 |
| 21 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
0.360 |
0.404 |
| 22 |
250372_at |
AT5G11460
|
unknown |
0.323 |
0.218 |
| 23 |
248738_at |
AT5G48020
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.304 |
0.048 |
| 24 |
264116_at |
AT2G31320
|
PARP1, POLY(ADP-RIBOSE) POLYMERASE 1 |
0.300 |
0.105 |
| 25 |
265449_at |
AT2G46610
|
At-RS31a, arginine/serine-rich splicing factor 31a, RS31a, arginine/serine-rich splicing factor 31a |
0.295 |
0.127 |
| 26 |
265184_at |
AT1G23710
|
unknown |
0.291 |
-0.017 |
| 27 |
248641_at |
AT5G49110
|
unknown |
0.282 |
0.036 |
| 28 |
254287_at |
AT4G22960
|
unknown |
0.281 |
0.213 |
| 29 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
0.274 |
-0.461 |
| 30 |
266063_at |
AT2G18760
|
CHR8, chromatin remodeling 8 |
0.221 |
0.039 |
| 31 |
264674_at |
AT1G09815
|
POLD4, polymerase delta 4 |
0.218 |
0.164 |
| 32 |
257086_at |
AT3G20490
|
unknown protein; Has 754 Blast hits to 165 proteins in 64 species: Archae - 0; Bacteria - 48; Metazoa - 26; Fungi - 25; Plants - 36; Viruses - 0; Other Eukaryotes - 619 (source: NCBI BLink). |
0.209 |
0.265 |
| 33 |
255812_at |
AT4G10310
|
ATHKT1, HKT1, high-affinity K+ transporter 1, HKT1;1 |
0.200 |
-0.546 |
| 34 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
0.194 |
-0.108 |
| 35 |
261953_at |
AT1G64440
|
REB1, ROOT EPIDERMAL BULGER1, RHD1, ROOT HAIR DEFECTIVE 1, UGE4, UDP-GLUCOSE 4-EPIMERASE |
0.189 |
0.111 |
| 36 |
261031_at |
AT1G17360
|
unknown |
0.189 |
-0.092 |
| 37 |
253514_at |
AT4G31805
|
POLAR, POLAR LOCALIZATION DURING ASYMMETRIC DIVISION AND REDISTRIBUTION |
0.186 |
0.002 |
| 38 |
258663_at |
AT3G08670
|
unknown |
0.185 |
0.222 |
| 39 |
263882_at |
AT2G21790
|
ATRNR1, RIBONUCLEOTIDE REDUCTASE LARGE SUBUNIT 1, CLS8, CRINKLY LEAVES 8, DPD2, defective in pollen organelle DNA degradation 2, R1, RIBONUCLEOTIDE REDUCTASE 1, RNR1, ribonucleotide reductase 1 |
0.181 |
0.440 |
| 40 |
267282_at |
AT2G19390
|
unknown |
0.175 |
0.139 |
| 41 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.270 |
-0.072 |
| 42 |
246072_at |
AT5G20240
|
PI, PISTILLATA |
-0.245 |
0.080 |
| 43 |
245385_at |
AT4G14020
|
[Rapid alkalinization factor (RALF) family protein] |
-0.240 |
-0.661 |
| 44 |
250683_x_at |
AT5G06640
|
EXT10, extensin 10 |
-0.169 |
-0.544 |
| 45 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.166 |
-0.031 |
| 46 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
-0.164 |
-0.399 |
| 47 |
255080_at |
AT4G09030
|
AGP10, arabinogalactan protein 10, ATAGP10 |
-0.141 |
-0.099 |
| 48 |
265175_at |
AT1G23480
|
ATCSLA03, cellulose synthase-like A3, ATCSLA3, CSLA03, CSLA03, cellulose synthase-like A3, CSLA3, CELLULOSE SYNTHASE-LIKE A3 |
-0.137 |
-0.295 |
| 49 |
264953_at |
AT1G77120
|
ADH, ALCOHOL DEHYDROGENASE, ADH1, alcohol dehydrogenase 1, ATADH, ARABIDOPSIS THALIANA ALCOHOL DEHYDROGENASE, ATADH1 |
-0.132 |
-0.267 |
| 50 |
263438_at |
AT2G28660
|
[Chloroplast-targeted copper chaperone protein] |
-0.130 |
-0.364 |
| 51 |
250301_at |
AT5G11970
|
unknown |
-0.126 |
-0.450 |
| 52 |
260812_at |
AT1G43650
|
UMAMIT22, Usually multiple acids move in and out Transporters 22 |
-0.124 |
-0.542 |
| 53 |
252624_at |
AT3G44735
|
ATPSK3, PHYTOSULFOKINE 3 PRECURSOR, PSK1, PSK3, PHYTOSULFOKINE 3 PRECURSOR |
-0.121 |
-0.582 |
| 54 |
255527_at |
AT4G02360
|
unknown |
-0.116 |
-0.033 |
| 55 |
259850_at |
AT1G72240
|
unknown |
-0.112 |
-0.537 |
| 56 |
259723_at |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
-0.110 |
-0.022 |
| 57 |
253140_at |
AT4G35480
|
RHA3B, RING-H2 finger A3B |
-0.109 |
-0.299 |
| 58 |
260569_at |
AT2G43640
|
[Signal recognition particle, SRP9/SRP14 subunit] |
-0.108 |
0.022 |
| 59 |
260557_at |
AT2G43610
|
[Chitinase family protein] |
-0.108 |
0.097 |
| 60 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.108 |
0.143 |
| 61 |
265356_at |
AT2G16595
|
[Translocon-associated protein (TRAP), alpha subunit] |
-0.107 |
-0.282 |
| 62 |
261209_at |
AT1G12810
|
[proline-rich family protein] |
-0.105 |
-0.440 |
| 63 |
259743_at |
AT1G71140
|
[MATE efflux family protein] |
-0.101 |
0.353 |
| 64 |
253329_at |
AT4G33480
|
unknown |
-0.101 |
-0.012 |
| 65 |
266094_at |
AT2G37975
|
[Yos1-like protein] |
-0.099 |
-0.075 |
| 66 |
258184_at |
AT3G21510
|
AHP1, histidine-containing phosphotransmitter 1 |
-0.098 |
-0.648 |
| 67 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.096 |
-0.268 |
| 68 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.093 |
-0.159 |
| 69 |
265884_at |
AT2G42320
|
[nucleolar protein gar2-related] |
-0.092 |
-0.309 |
| 70 |
247199_at |
AT5G65210
|
TGA1, TGACG sequence-specific binding protein 1 |
-0.091 |
-0.075 |
| 71 |
264815_at |
AT1G03620
|
[ELMO/CED-12 family protein] |
-0.091 |
-0.072 |
| 72 |
258935_at |
AT3G10120
|
unknown |
-0.091 |
0.143 |
| 73 |
246258_at |
AT1G31840
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.091 |
0.162 |
| 74 |
246485_at |
AT5G16080
|
AtCXE17, carboxyesterase 17, CXE17, carboxyesterase 17 |
-0.091 |
0.016 |
| 75 |
264938_at |
AT1G60610
|
[SBP (S-ribonuclease binding protein) family protein] |
-0.090 |
-0.184 |
| 76 |
260355_at |
AT1G69180
|
CRC, CRABS CLAW |
-0.090 |
-0.009 |
| 77 |
257029_at |
AT3G19240
|
[Vacuolar import/degradation, Vid27-related protein] |
-0.087 |
0.132 |
| 78 |
260849_at |
AT1G21860
|
sks7, SKU5 similar 7 |
-0.086 |
-0.015 |
| 79 |
260700_at |
AT1G32260
|
unknown |
-0.084 |
-0.171 |