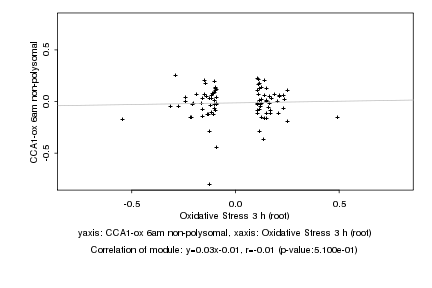
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Oxidative Stress 3 h (root) |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
257670_at |
AT3G20340
|
unknown |
0.490 |
-0.153 |
| 2 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.249 |
-0.185 |
| 3 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
0.247 |
0.116 |
| 4 |
254313_at |
AT4G22460
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.236 |
0.020 |
| 5 |
267519_at |
AT2G30470
|
HSI2, high-level expression of sugar-inducible gene 2, VAL1, VP1/ABI3-LIKE 1 |
0.229 |
-0.067 |
| 6 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
0.228 |
0.064 |
| 7 |
245584_at |
AT4G14940
|
AO1, amine oxidase 1, ATAO1, amine oxidase 1 |
0.212 |
0.051 |
| 8 |
249230_at |
AT5G42070
|
unknown |
0.211 |
0.060 |
| 9 |
250204_at |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
0.205 |
-0.115 |
| 10 |
257479_at |
AT1G16150
|
WAKL4, wall associated kinase-like 4 |
0.200 |
0.002 |
| 11 |
247912_at |
AT5G57480
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.187 |
0.068 |
| 12 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
0.172 |
0.035 |
| 13 |
258143_at |
AT3G18170
|
[Glycosyltransferase family 61 protein] |
0.169 |
-0.086 |
| 14 |
266055_at |
AT2G40650
|
[PRP38 family protein] |
0.167 |
-0.112 |
| 15 |
256713_at |
AT2G34060
|
[Peroxidase superfamily protein] |
0.163 |
-0.011 |
| 16 |
263957_at |
AT2G35880
|
[TPX2 (targeting protein for Xklp2) protein family] |
0.160 |
0.049 |
| 17 |
267418_at |
AT2G35000
|
ATL9, Arabidopsis toxicos en levadura 9 |
0.159 |
-0.056 |
| 18 |
267635_at |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
0.148 |
-0.109 |
| 19 |
262309_at |
AT1G70820
|
[phosphoglucomutase, putative / glucose phosphomutase, putative] |
0.148 |
0.012 |
| 20 |
252439_at |
AT3G47400
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
0.147 |
-0.157 |
| 21 |
253790_at |
AT4G28660
|
PSB28, photosystem II reaction center PSB28 protein |
0.146 |
0.003 |
| 22 |
253919_at |
AT4G27350
|
unknown |
0.145 |
0.132 |
| 23 |
266011_at |
AT2G37440
|
[DNAse I-like superfamily protein] |
0.140 |
0.066 |
| 24 |
265643_at |
AT2G27390
|
[proline-rich family protein] |
0.137 |
0.209 |
| 25 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.136 |
-0.164 |
| 26 |
266920_at |
AT2G45750
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.135 |
-0.361 |
| 27 |
266222_at |
AT2G28780
|
unknown |
0.125 |
0.023 |
| 28 |
263222_at |
AT1G30640
|
[Protein kinase family protein] |
0.123 |
0.142 |
| 29 |
265852_at |
AT2G42350
|
[RING/U-box superfamily protein] |
0.122 |
-0.147 |
| 30 |
251061_at |
AT5G01830
|
SAUR21, SMALL AUXIN UP RNA 21 |
0.121 |
-0.010 |
| 31 |
250095_at |
AT5G17230
|
PSY, PHYTOENE SYNTHASE |
0.120 |
-0.043 |
| 32 |
251540_at |
AT3G58740
|
CSY1, citrate synthase 1 |
0.118 |
-0.028 |
| 33 |
253425_at |
AT4G32190
|
[Myosin heavy chain-related protein] |
0.115 |
0.128 |
| 34 |
250366_at |
AT5G11420
|
unknown |
0.115 |
0.012 |
| 35 |
262502_at |
AT1G21600
|
PTAC6, plastid transcriptionally active 6 |
0.115 |
0.184 |
| 36 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
0.114 |
-0.288 |
| 37 |
266207_at |
AT2G27680
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.114 |
-0.077 |
| 38 |
264696_at |
AT1G70230
|
AXY4, ALTERED XYLOGLUCAN 4, TBL27, TRICHOME BIREFRINGENCE-LIKE 27 |
0.110 |
0.219 |
| 39 |
250661_at |
AT5G07030
|
[Eukaryotic aspartyl protease family protein] |
0.108 |
0.073 |
| 40 |
267209_at |
AT2G30930
|
unknown |
0.107 |
0.171 |
| 41 |
262444_at |
AT1G47480
|
[alpha/beta-Hydrolases superfamily protein] |
0.106 |
-0.016 |
| 42 |
260704_at |
AT1G32470
|
[Single hybrid motif superfamily protein] |
0.106 |
-0.081 |
| 43 |
250376_at |
AT5G11550
|
[ARM repeat superfamily protein] |
0.104 |
0.232 |
| 44 |
258149_at |
AT3G18110
|
EMB1270, embryo defective 1270 |
0.104 |
-0.024 |
| 45 |
258167_at |
AT3G21560
|
BRT1, Bright Trichomes 1, UGT84A2, UDP-glucosyl transferase 84A2 |
0.103 |
-0.114 |
| 46 |
264559_at |
AT1G09610
|
GXM3, glucuronoxylan methyltransferase 3 |
0.103 |
0.113 |
| 47 |
249333_at |
AT5G40990
|
GLIP1, GDSL lipase 1 |
-0.548 |
-0.167 |
| 48 |
260394_at |
AT1G74080
|
ATMYB122, MYB DOMAIN PROTEIN 122, MYB122, myb domain protein 122 |
-0.317 |
-0.040 |
| 49 |
266821_at |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
-0.290 |
0.253 |
| 50 |
258606_at |
AT3G02840
|
[ARM repeat superfamily protein] |
-0.279 |
-0.045 |
| 51 |
266292_at |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-0.245 |
0.047 |
| 52 |
246145_at |
AT5G19880
|
[Peroxidase superfamily protein] |
-0.243 |
0.008 |
| 53 |
249188_at |
AT5G42830
|
[HXXXD-type acyl-transferase family protein] |
-0.220 |
-0.150 |
| 54 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
-0.216 |
-0.149 |
| 55 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-0.210 |
-0.027 |
| 56 |
256759_at |
AT3G25640
|
unknown |
-0.206 |
-0.013 |
| 57 |
253044_at |
AT4G37290
|
unknown |
-0.188 |
0.070 |
| 58 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
-0.164 |
-0.018 |
| 59 |
245375_at |
AT4G17660
|
[Protein kinase superfamily protein] |
-0.162 |
-0.143 |
| 60 |
249459_at |
AT5G39580
|
[Peroxidase superfamily protein] |
-0.159 |
0.033 |
| 61 |
245875_at |
AT1G26240
|
[Proline-rich extensin-like family protein] |
-0.159 |
-0.075 |
| 62 |
253182_at |
AT4G35190
|
LOG5, LONELY GUY 5 |
-0.154 |
0.204 |
| 63 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.151 |
0.074 |
| 64 |
249001_at |
AT5G44990
|
[Glutathione S-transferase family protein] |
-0.149 |
0.176 |
| 65 |
257061_at |
AT3G18250
|
[Putative membrane lipoprotein] |
-0.141 |
0.051 |
| 66 |
252539_at |
AT3G45730
|
unknown |
-0.138 |
-0.125 |
| 67 |
249770_at |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
-0.133 |
-0.119 |
| 68 |
266197_at |
AT2G39120
|
WTF9, what's this factor 9 |
-0.129 |
-0.794 |
| 69 |
257026_at |
AT3G19200
|
unknown |
-0.127 |
0.035 |
| 70 |
265960_at |
AT2G37470
|
[Histone superfamily protein] |
-0.127 |
-0.281 |
| 71 |
245777_at |
AT1G73540
|
atnudt21, nudix hydrolase homolog 21, NUDT21, nudix hydrolase homolog 21 |
-0.125 |
-0.035 |
| 72 |
246639_x_at |
AT5G34895
|
[similar to heat shock protein binding [Arabidopsis thaliana] (TAIR:AT1G32830.1)] |
-0.118 |
0.067 |
| 73 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
-0.118 |
-0.102 |
| 74 |
260278_at |
AT1G80590
|
ATWRKY66, WRKY66, WRKY DNA-binding protein 66 |
-0.116 |
0.030 |
| 75 |
250287_at |
AT5G13330
|
Rap2.6L, related to AP2 6l |
-0.112 |
0.087 |
| 76 |
266616_at |
AT2G29680
|
ATCDC6, CDC6, cell division control 6 |
-0.109 |
-0.122 |
| 77 |
255284_at |
AT4G04610
|
APR, APR1, APS reductase 1, ATAPR1, PRH19, PAPS REDUCTASE HOMOLOG 19 |
-0.107 |
0.084 |
| 78 |
254625_at |
AT4G18470
|
SNI1, SUPPRESSOR OF NPR1-1, INDUCIBLE 1 |
-0.103 |
0.196 |
| 79 |
253795_at |
AT4G28420
|
[Tyrosine transaminase family protein] |
-0.102 |
0.092 |
| 80 |
261518_at |
AT1G71695
|
[Peroxidase superfamily protein] |
-0.101 |
-0.021 |
| 81 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
-0.101 |
0.013 |
| 82 |
253915_at |
AT4G27280
|
[Calcium-binding EF-hand family protein] |
-0.101 |
-0.058 |
| 83 |
252251_at |
AT3G49820
|
unknown |
-0.100 |
0.109 |
| 84 |
252506_at |
AT3G46180
|
ATUTR5, UDP-GALACTOSE TRANSPORTER 5, UTR5, UDP-galactose transporter 5 |
-0.099 |
0.137 |
| 85 |
256306_at |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
-0.098 |
0.134 |
| 86 |
265742_at |
AT2G01290
|
RPI2, ribose-5-phosphate isomerase 2 |
-0.098 |
-0.084 |
| 87 |
249751_at |
AT5G24650
|
[Mitochondrial import inner membrane translocase subunit Tim17/Tim22/Tim23 family protein] |
-0.096 |
-0.441 |
| 88 |
264024_at |
AT2G21180
|
unknown |
-0.096 |
-0.027 |
| 89 |
247625_at |
AT5G60200
|
TMO6, TARGET OF MONOPTEROS 6 |
-0.094 |
-0.011 |
| 90 |
245855_at |
AT5G13550
|
SULTR4;1, sulfate transporter 4.1 |
-0.092 |
0.126 |
| 91 |
249771_at |
AT5G24080
|
[Protein kinase superfamily protein] |
-0.092 |
0.045 |