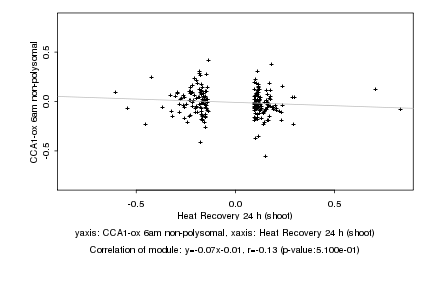
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Heat Recovery 24 h (shoot) |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
0.830 |
-0.079 |
| 2 |
249575_at |
AT5G37670
|
[HSP20-like chaperones superfamily protein] |
0.703 |
0.123 |
| 3 |
251720_at |
AT3G56160
|
[Sodium Bile acid symporter family] |
0.293 |
0.047 |
| 4 |
265675_at |
AT2G32120
|
HSP70T-2, heat-shock protein 70T-2 |
0.289 |
-0.229 |
| 5 |
248330_at |
AT5G52810
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.284 |
0.047 |
| 6 |
246284_at |
AT4G36780
|
BEH2, BES1/BZR1 homolog 2 |
0.237 |
-0.032 |
| 7 |
246944_at |
AT5G25450
|
[Cytochrome bd ubiquinol oxidase, 14kDa subunit] |
0.234 |
0.158 |
| 8 |
260693_at |
AT1G32450
|
AtNPF7.3, NPF7.3, NRT1/ PTR family 7.3, NRT1.5, nitrate transporter 1.5 |
0.230 |
-0.188 |
| 9 |
249740_at |
AT5G24500
|
unknown |
0.228 |
-0.105 |
| 10 |
244903_at |
ATMG00660
|
ORF149 |
0.219 |
-0.091 |
| 11 |
259661_at |
AT1G55265
|
unknown |
0.204 |
-0.084 |
| 12 |
257066_at |
AT3G18280
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.203 |
-0.070 |
| 13 |
261317_at |
AT1G53030
|
[Cytochrome C oxidase copper chaperone (COX17)] |
0.198 |
-0.036 |
| 14 |
259100_at |
AT3G04880
|
DRT102, DNA-DAMAGE-REPAIR/TOLERATION 2 |
0.191 |
-0.075 |
| 15 |
255777_at |
AT1G18630
|
GR-RBP6, glycine-rich RNA-binding protein 6 |
0.190 |
-0.059 |
| 16 |
260025_at |
AT1G30070
|
[SGS domain-containing protein] |
0.182 |
-0.055 |
| 17 |
265142_at |
AT1G51360
|
ATDABB1, DIMERIC A/B BARREL DOMAINS-PROTEIN 1, DABB1, dimeric A/B barrel domainS-protein 1 |
0.178 |
0.382 |
| 18 |
262098_at |
AT1G56170
|
ATHAP5B, HAP5B, NF-YC2, nuclear factor Y, subunit C2 |
0.176 |
0.112 |
| 19 |
247642_at |
AT5G60590
|
[DHBP synthase RibB-like alpha/beta domain] |
0.173 |
0.055 |
| 20 |
247434_at |
AT5G62575
|
SDH7, succinate dehydrogenase 7, SDH7B, succinate dehydrogenase 7B |
0.173 |
-0.043 |
| 21 |
255659_at |
AT4G00895
|
[ATPase, F1 complex, OSCP/delta subunit protein] |
0.172 |
0.030 |
| 22 |
253058_at |
AT4G37660
|
[Ribosomal protein L12/ ATP-dependent Clp protease adaptor protein ClpS family protein] |
0.171 |
-0.145 |
| 23 |
259056_at |
AT3G03420
|
[Ku70-binding family protein] |
0.168 |
0.191 |
| 24 |
247497_at |
AT5G61770
|
PPAN, PETER PAN-like protein |
0.167 |
0.037 |
| 25 |
244943_at |
ATMG00070
|
NAD9, NADH dehydrogenase subunit 9 |
0.165 |
-0.183 |
| 26 |
254079_at |
AT4G25730
|
[FtsJ-like methyltransferase family protein] |
0.164 |
-0.010 |
| 27 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.163 |
-0.031 |
| 28 |
254169_at |
AT4G24290
|
[MAC/Perforin domain-containing protein] |
0.161 |
0.080 |
| 29 |
266054_at |
AT2G40640
|
[RING/U-box superfamily protein] |
0.159 |
-0.030 |
| 30 |
252981_at |
AT4G38260
|
unknown |
0.157 |
-0.001 |
| 31 |
248429_at |
AT5G51770
|
[Protein kinase superfamily protein] |
0.157 |
-0.191 |
| 32 |
257791_at |
AT3G27110
|
[Peptidase family M48 family protein] |
0.157 |
-0.055 |
| 33 |
250477_at |
AT5G10190
|
[Major facilitator superfamily protein] |
0.155 |
-0.074 |
| 34 |
246612_at |
AT5G35320
|
unknown |
0.155 |
0.016 |
| 35 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.152 |
0.114 |
| 36 |
266872_at |
AT2G44730
|
[Alcohol dehydrogenase transcription factor Myb/SANT-like family protein] |
0.148 |
-0.554 |
| 37 |
246914_at |
AT5G25860
|
[F-box/RNI-like superfamily protein] |
0.147 |
-0.072 |
| 38 |
255412_at |
AT4G02980
|
ABP, ABP1, endoplasmic reticulum auxin binding protein 1 |
0.146 |
-0.008 |
| 39 |
249606_at |
AT5G37260
|
CIR1, CIRCADIAN 1, RVE2, REVEILLE 2 |
0.146 |
-0.209 |
| 40 |
250304_at |
AT5G12110
|
[Glutathione S-transferase, C-terminal-like] |
0.144 |
-0.097 |
| 41 |
254878_at |
AT4G11660
|
AT-HSFB2B, HSF7, HSFB2B, HEAT SHOCK TRANSCRIPTION FACTOR B2B |
0.141 |
-0.227 |
| 42 |
256522_at |
AT1G66160
|
ATCMPG1, CMPG1, CYS, MET, PRO, and GLY protein 1 |
0.139 |
-0.106 |
| 43 |
258998_at |
AT3G01820
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.138 |
-0.115 |
| 44 |
254707_at |
AT4G18010
|
5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, AT5PTASE2, myo-inositol polyphosphate 5-phosphatase 2, IP5PII, INOSITOL(1,4,5)P3 5-PHOSPHATASE II |
0.131 |
-0.063 |
| 45 |
259130_at |
AT3G02190
|
[Ribosomal protein L39 family protein] |
0.130 |
-0.164 |
| 46 |
247653_at |
AT5G59950
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.130 |
-0.041 |
| 47 |
262295_at |
AT1G27650
|
ATU2AF35A |
0.125 |
0.041 |
| 48 |
262625_at |
AT1G06440
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
0.123 |
-0.072 |
| 49 |
256333_at |
AT1G76860
|
LSM3B, SM-like 3B |
0.122 |
-0.087 |
| 50 |
264867_at |
AT1G24150
|
ATFH4, FORMIN HOMOLOGUE 4, FH4, formin homologue 4 |
0.122 |
-0.026 |
| 51 |
252982_at |
AT4G38130
|
ATHD1, ARABIDOPSIS HISTONE DEACETYLASE 1, ATHDA19, ARABIDOPSIS HISTONE DEACETYLASE 19, HD1, histone deacetylase 1, HDA1, HDA19, HISTONE DEACETYLASE 19, RPD3A |
0.119 |
0.024 |
| 52 |
252229_at |
AT3G49890
|
unknown |
0.119 |
-0.087 |
| 53 |
253865_at |
AT4G27470
|
ATRMA3, RMA3, RING membrane-anchor 3 |
0.118 |
-0.115 |
| 54 |
264013_at |
AT2G21070
|
FIO1, FIONA1 |
0.118 |
0.124 |
| 55 |
260054_at |
AT1G78130
|
UNE2, unfertilized embryo sac 2 |
0.118 |
-0.017 |
| 56 |
265030_at |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
0.117 |
0.019 |
| 57 |
265258_at |
AT2G20390
|
unknown |
0.117 |
0.030 |
| 58 |
250495_at |
AT5G09770
|
[Ribosomal protein L17 family protein] |
0.116 |
-0.060 |
| 59 |
253603_at |
AT4G30935
|
ATWRKY32, WRKY32, WRKY DNA-binding protein 32 |
0.116 |
-0.346 |
| 60 |
262840_at |
AT1G14900
|
HMGA, high mobility group A |
0.115 |
0.005 |
| 61 |
261260_at |
AT1G26680
|
[transcriptional factor B3 family protein] |
0.115 |
0.053 |
| 62 |
249930_at |
AT5G22360
|
ATVAMP714, vesicle-associated membrane protein 714, VAMP714, VAMP714, vesicle-associated membrane protein 714 |
0.114 |
0.093 |
| 63 |
266687_at |
AT2G19670
|
ATPRMT1A, ARABIDOPSIS THALIANA PROTEIN ARGININE METHYLTRANSFERASE 1A, PRMT1A, protein arginine methyltransferase 1A |
0.114 |
-0.042 |
| 64 |
254251_at |
AT4G23300
|
CRK22, cysteine-rich RLK (RECEPTOR-like protein kinase) 22 |
0.114 |
0.177 |
| 65 |
253917_at |
AT4G27380
|
unknown |
0.113 |
0.161 |
| 66 |
254778_at |
AT4G12750
|
[Homeodomain-like transcriptional regulator] |
0.112 |
0.146 |
| 67 |
257483_at |
AT1G49620
|
AtKRP7, ICK5, ICN6, KRP7, KIP-RELATED PROTEIN 7 |
0.111 |
0.112 |
| 68 |
256372_at |
AT1G66750
|
AT;CDKD;2, CAK4, CDK-activating kinase 4, CAK4AT, CDK-ACTIVATING KINASE 4, CDKD1;2, CYCLIN-DEPENDENT KINASE D1;2, CDKD;2 |
0.111 |
0.076 |
| 69 |
261762_at |
AT1G15510
|
ATECB2, ARABIDOPSIS EARLY CHLOROPLAST BIOGENESIS2, ECB2, EARLY CHLOROPLAST BIOGENESIS2, VAC1, VANILLA CREAM 1 |
0.111 |
-0.034 |
| 70 |
256235_at |
AT3G12490
|
ATCYS6, ARABIDOPSIS THALIANA PHYTOCYSTATIN 6, ATCYSB, cystatin B, CYSB, cystatin B |
0.111 |
0.304 |
| 71 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.110 |
-0.127 |
| 72 |
257435_at |
AT2G24590
|
At-RSZ22a, RS-containing zinc finger protein 22a, RSZ22a, RS-containing zinc finger protein 22a |
0.110 |
0.024 |
| 73 |
253878_at |
AT4G27550
|
ATTPS4, ARABIDOPSIS THALIANA TREHALOSE PHOSPHATASE/SYNTHASE 4, TPS4, trehalose-6-phosphatase synthase S4 |
0.110 |
0.016 |
| 74 |
245463_at |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
0.109 |
-0.158 |
| 75 |
256353_at |
AT1G55000
|
[peptidoglycan-binding LysM domain-containing protein] |
0.109 |
-0.181 |
| 76 |
258162_at |
AT3G17810
|
PYD1, pyrimidine 1 |
0.109 |
0.101 |
| 77 |
266622_at |
AT2G35430
|
[Zinc finger C-x8-C-x5-C-x3-H type family protein] |
0.107 |
-0.027 |
| 78 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
0.105 |
-0.071 |
| 79 |
250470_at |
AT5G10160
|
[Thioesterase superfamily protein] |
0.104 |
-0.029 |
| 80 |
266635_at |
AT2G35470
|
unknown |
0.104 |
0.184 |
| 81 |
266118_at |
AT2G02130
|
LCR68, low-molecular-weight cysteine-rich 68, PDF2.3 |
0.103 |
0.084 |
| 82 |
250886_at |
AT5G04440
|
unknown |
0.102 |
-0.062 |
| 83 |
250485_at |
AT5G09990
|
PROPEP5, elicitor peptide 5 precursor |
0.102 |
0.090 |
| 84 |
251283_at |
AT3G61790
|
[Protein with RING/U-box and TRAF-like domains] |
0.102 |
0.004 |
| 85 |
250707_at |
AT5G05950
|
MEE60, maternal effect embryo arrest 60 |
0.101 |
0.231 |
| 86 |
262113_at |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
0.101 |
-0.026 |
| 87 |
255053_at |
AT4G09730
|
RH39, RH39 |
0.100 |
-0.112 |
| 88 |
264865_at |
AT1G24120
|
ARL1, ARG1-like 1 |
0.100 |
-0.367 |
| 89 |
257457_at |
AT2G05430
|
[Ubiquitin-specific protease family C19-related protein] |
0.099 |
0.023 |
| 90 |
252273_at |
AT3G49660
|
AtWDR5a, WDR5a, human WDR5 (WD40 repeat) homolog a |
0.099 |
-0.021 |
| 91 |
262818_at |
AT1G11755
|
LEW1, LEAF WILTING 1 |
0.098 |
-0.169 |
| 92 |
255887_at |
AT1G20370
|
[Pseudouridine synthase family protein] |
0.098 |
0.027 |
| 93 |
252667_at |
AT3G44200
|
ATNEK6, NIMA-RELATED KINASE6, IBO1, NEK6, NIMA (never in mitosis, gene A)-related 6 |
0.098 |
0.051 |
| 94 |
264548_at |
AT1G55680
|
[Transducin/WD40 repeat-like superfamily protein] |
0.097 |
-0.050 |
| 95 |
246966_at |
AT5G24850
|
CRY3, cryptochrome 3 |
0.097 |
0.068 |
| 96 |
262775_at |
AT1G13000
|
unknown |
0.097 |
-0.014 |
| 97 |
259887_at |
AT1G76360
|
[Protein kinase superfamily protein] |
0.096 |
-0.048 |
| 98 |
256086_at |
AT1G20770
|
unknown |
0.096 |
-0.060 |
| 99 |
245747_at |
AT1G51100
|
CRR41, CHLORORESPIRATORY REDUCTION 41 |
0.096 |
-0.086 |
| 100 |
252498_at |
AT3G46810
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.095 |
-0.010 |
| 101 |
255017_at |
AT4G10100
|
CNX7, co-factor for nitrate, reductase and xanthine dehydrogenase 7, SIR5 |
0.095 |
-0.154 |
| 102 |
266796_at |
AT2G02880
|
[mucin-related] |
0.095 |
-0.003 |
| 103 |
257001_at |
AT3G14080
|
LSM1B, SM-like 1B |
0.094 |
0.195 |
| 104 |
266315_at |
AT2G27100
|
SE, SERRATE |
0.094 |
-0.023 |
| 105 |
258996_at |
AT3G01800
|
[Ribosome recycling factor] |
0.094 |
-0.063 |
| 106 |
245303_at |
AT4G17010
|
unknown |
0.093 |
-0.182 |
| 107 |
265043_at |
AT1G03900
|
ABCI18, ATP-binding cassette I18, ATNAP4, non-intrinsic ABC protein 4, NAP4, non-intrinsic ABC protein 4 |
0.093 |
0.023 |
| 108 |
257733_at |
AT3G18350
|
unknown |
0.092 |
0.053 |
| 109 |
257093_at |
AT3G20570
|
AtENODL9, ENODL9, early nodulin-like protein 9 |
0.092 |
0.127 |
| 110 |
267545_at |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
-0.609 |
0.101 |
| 111 |
256916_at |
AT3G24050
|
GATA1, GATA transcription factor 1 |
-0.549 |
-0.065 |
| 112 |
252073_at |
AT3G51750
|
unknown |
-0.457 |
-0.227 |
| 113 |
251179_at |
AT3G63460
|
SEC31B |
-0.424 |
0.248 |
| 114 |
253243_at |
AT4G34560
|
unknown |
-0.370 |
-0.052 |
| 115 |
258733_at |
AT3G05850
|
MUG7, MUSTANG 7 |
-0.329 |
0.064 |
| 116 |
253991_at |
AT4G26000
|
PEP, PEPPER |
-0.326 |
-0.100 |
| 117 |
260395_at |
AT1G69780
|
ATHB13 |
-0.322 |
-0.148 |
| 118 |
264954_at |
AT1G77060
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.304 |
0.053 |
| 119 |
260126_at |
AT1G36370
|
SHM7, serine hydroxymethyltransferase 7 |
-0.297 |
0.095 |
| 120 |
265387_at |
AT2G20670
|
unknown |
-0.293 |
0.083 |
| 121 |
258206_at |
AT3G14010
|
CID4, CTC-interacting domain 4 |
-0.285 |
-0.029 |
| 122 |
250446_at |
AT5G10770
|
[Eukaryotic aspartyl protease family protein] |
-0.283 |
-0.108 |
| 123 |
259617_at |
AT1G47970
|
unknown |
-0.278 |
0.024 |
| 124 |
249168_at |
AT5G42870
|
ATPAH2, PHOSPHATIDIC ACID PHOSPHOHYDROLASE 2, PAH2, phosphatidic acid phosphohydrolase 2 |
-0.274 |
0.039 |
| 125 |
252957_at |
AT4G38680
|
ATCSP2, ARABIDOPSIS THALIANA COLD SHOCK PROTEIN 2, CSDP2, COLD SHOCK DOMAIN PROTEIN 2, CSP2, COLD SHOCK PROTEIN 2, GRP2, glycine rich protein 2 |
-0.266 |
0.066 |
| 126 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.262 |
-0.036 |
| 127 |
247056_at |
AT5G66750
|
ATDDM1, CHA1, CHR01, CHROMATIN REMODELING 1, CHR1, chromatin remodeling 1, DDM1, DECREASED DNA METHYLATION 1, SOM1, SOMNIFEROUS 1, SOM4 |
-0.261 |
-0.052 |
| 128 |
263829_at |
AT2G40435
|
unknown |
-0.260 |
-0.167 |
| 129 |
262801_at |
AT1G21010
|
unknown |
-0.258 |
0.045 |
| 130 |
258097_at |
AT3G23660
|
[Sec23/Sec24 protein transport family protein] |
-0.249 |
-0.028 |
| 131 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.243 |
-0.204 |
| 132 |
266361_at |
AT2G32450
|
[Calcium-binding tetratricopeptide family protein] |
-0.236 |
0.012 |
| 133 |
261382_at |
AT1G05470
|
CVP2, COTYLEDON VASCULAR PATTERN 2 |
-0.233 |
-0.143 |
| 134 |
247828_at |
AT5G58510
|
unknown |
-0.233 |
0.111 |
| 135 |
253323_at |
AT4G33920
|
APD5, Arabidopsis Pp2c clade D 5 |
-0.230 |
0.151 |
| 136 |
258225_at |
AT3G15630
|
unknown |
-0.229 |
0.105 |
| 137 |
264771_at |
AT1G22940
|
TH-1, TH1, THIAMINE REQUIRING 1, THIE, THIAMINEE |
-0.229 |
0.084 |
| 138 |
255506_at |
AT4G02130
|
GATL6, galacturonosyltransferase-like 6, LGT10 |
-0.228 |
0.014 |
| 139 |
249577_at |
AT5G37710
|
[alpha/beta-Hydrolases superfamily protein] |
-0.228 |
-0.134 |
| 140 |
254966_at |
AT4G11110
|
SPA2, SPA1-related 2 |
-0.223 |
0.083 |
| 141 |
247530_at |
AT5G61540
|
[N-terminal nucleophile aminohydrolases (Ntn hydrolases) superfamily protein] |
-0.221 |
0.164 |
| 142 |
263599_at |
AT2G01830
|
AHK4, ARABIDOPSIS HISTIDINE KINASE 4, ATCRE1, CRE1, CYTOKININ RESPONSE 1, WOL, WOODEN LEG, WOL1, WOODEN LEG 1 |
-0.219 |
0.098 |
| 143 |
264264_at |
AT1G09250
|
AIF4, ATBS1 Interacting Factor 4 |
-0.217 |
-0.009 |
| 144 |
265875_at |
AT2G01690
|
[ARM repeat superfamily protein] |
-0.217 |
-0.048 |
| 145 |
246801_at |
AT5G26830
|
[Threonyl-tRNA synthetase] |
-0.215 |
0.020 |
| 146 |
251688_at |
AT3G56480
|
[myosin heavy chain-related] |
-0.210 |
0.065 |
| 147 |
265265_at |
AT2G42900
|
[Plant basic secretory protein (BSP) family protein] |
-0.209 |
0.236 |
| 148 |
260613_at |
AT1G53380
|
unknown |
-0.205 |
-0.109 |
| 149 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.203 |
-0.070 |
| 150 |
249370_at |
AT5G40710
|
[zinc finger (C2H2 type) family protein] |
-0.201 |
0.036 |
| 151 |
255255_at |
AT4G05070
|
[Wound-responsive family protein] |
-0.201 |
-0.044 |
| 152 |
265844_at |
AT2G35620
|
FEI2, FEI 2 |
-0.198 |
0.216 |
| 153 |
264625_at |
AT1G09020
|
ATSNF4, SNF4, homolog of yeast sucrose nonfermenting 4 |
-0.197 |
-0.057 |
| 154 |
247955_at |
AT5G56950
|
NAP1;3, nucleosome assembly protein 1;3, NFA03, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A 03, NFA3, NUCLEOSOME/CHROMATIN ASSEMBLY FACTOR GROUP A3 |
-0.196 |
-0.102 |
| 155 |
264080_at |
AT2G28520
|
VHA-A1, vacuolar proton ATPase A1 |
-0.192 |
0.191 |
| 156 |
267330_at |
AT2G19270
|
unknown |
-0.188 |
0.044 |
| 157 |
264206_at |
AT1G22730
|
[MA3 domain-containing protein] |
-0.186 |
-0.029 |
| 158 |
258146_at |
AT3G18060
|
[transducin family protein / WD-40 repeat family protein] |
-0.184 |
0.044 |
| 159 |
259713_at |
AT1G77610
|
[EamA-like transporter family protein] |
-0.183 |
0.290 |
| 160 |
247845_at |
AT5G58090
|
[O-Glycosyl hydrolases family 17 protein] |
-0.182 |
0.306 |
| 161 |
259530_at |
AT1G12450
|
[SNARE associated Golgi protein family] |
-0.182 |
0.124 |
| 162 |
262909_at |
AT1G59830
|
PP2A-1, protein phosphatase 2A-1 |
-0.181 |
-0.406 |
| 163 |
261424_at |
AT1G18700
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.179 |
-0.060 |
| 164 |
246505_at |
AT5G16250
|
unknown |
-0.178 |
0.096 |
| 165 |
246542_at |
AT5G15020
|
SNL2, SIN3-like 2 |
-0.178 |
0.267 |
| 166 |
266650_at |
AT2G25800
|
unknown |
-0.176 |
-0.063 |
| 167 |
261796_at |
AT1G30440
|
[Phototropic-responsive NPH3 family protein] |
-0.176 |
0.119 |
| 168 |
263939_at |
AT2G36070
|
ATTIM44-2, translocase inner membrane subunit 44-2, TIM44-2, translocase inner membrane subunit 44-2 |
-0.176 |
0.174 |
| 169 |
266698_at |
AT2G19830
|
SNF7.2, VPS32 |
-0.175 |
-0.100 |
| 170 |
253448_at |
AT4G32640
|
[Sec23/Sec24 protein transport family protein] |
-0.175 |
0.082 |
| 171 |
256075_at |
AT1G18150
|
ATMPK8, MPK8 |
-0.174 |
-0.179 |
| 172 |
250034_at |
AT5G18280
|
APY2, apyrase 2, ATAPY2, apyrase 2 |
-0.173 |
0.076 |
| 173 |
249171_at |
AT5G42940
|
[RING/U-box superfamily protein] |
-0.173 |
-0.124 |
| 174 |
252502_at |
AT3G46900
|
COPT2, copper transporter 2 |
-0.173 |
0.109 |
| 175 |
258045_at |
AT3G21280
|
UBP7, ubiquitin-specific protease 7 |
-0.173 |
-0.011 |
| 176 |
258909_at |
AT3G06340
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.170 |
-0.134 |
| 177 |
252066_at |
AT3G51550
|
FER, FERONIA |
-0.170 |
0.023 |
| 178 |
264547_at |
AT1G55620
|
ATCLC-F, CLC-F, chloride channel F, CLCF, CHLORIDE CHANNEL F |
-0.170 |
-0.010 |
| 179 |
263654_at |
AT1G04300
|
[TRAF-like superfamily protein] |
-0.170 |
0.147 |
| 180 |
253406_at |
AT4G32890
|
GATA9, GATA transcription factor 9 |
-0.169 |
-0.148 |
| 181 |
248933_at |
AT5G46070
|
[Guanylate-binding family protein] |
-0.168 |
0.001 |
| 182 |
250617_at |
AT5G07290
|
AML4, MEI2-like 4, ML4, MEI2-like 4 |
-0.168 |
-0.014 |
| 183 |
254270_at |
AT4G23100
|
ATECS1, AtGSH1, CAD2, CADMIUM SENSITIVE 2, GSH1, glutamate-cysteine ligase, GSHA, PAD2, PHYTOALEXIN DEFICIENT 2, RML1, ROOT MERISTEMLESS 1 |
-0.168 |
-0.191 |
| 184 |
265467_at |
AT2G37050
|
[Leucine-rich repeat protein kinase family protein] |
-0.167 |
-0.173 |
| 185 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.166 |
0.091 |
| 186 |
265877_at |
AT2G42380
|
ATBZIP34, BZIP34 |
-0.166 |
0.088 |
| 187 |
246928_at |
AT5G25270
|
[Ubiquitin-like superfamily protein] |
-0.165 |
0.060 |
| 188 |
264442_at |
AT1G27480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.164 |
0.011 |
| 189 |
249337_at |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
-0.163 |
0.058 |
| 190 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.163 |
0.006 |
| 191 |
253753_at |
AT4G29030
|
[Putative membrane lipoprotein] |
-0.163 |
-0.015 |
| 192 |
255558_at |
AT4G01900
|
GLB1, GLNB1 homolog, PII |
-0.161 |
-0.026 |
| 193 |
257951_at |
AT3G21700
|
ATSGP2, SGP2 |
-0.160 |
0.046 |
| 194 |
250693_at |
AT5G06600
|
AtUBP12, UBP12, ubiquitin-specific protease 12 |
-0.158 |
-0.211 |
| 195 |
255832_at |
AT2G33360
|
unknown |
-0.157 |
0.014 |
| 196 |
261105_at |
AT1G63000
|
NRS/ER, nucleotide-rhamnose synthase/epimerase-reductase, UER1, UDP-4-KETO-6-DEOXY-D-GLUCOSE-3,5-EPIMERASE-4-REDUCTASE 1 |
-0.155 |
-0.258 |
| 197 |
258986_at |
AT3G08910
|
[DNAJ heat shock family protein] |
-0.155 |
-0.160 |
| 198 |
252292_at |
AT3G49000
|
[RNA polymerase III subunit RPC82 family protein] |
-0.155 |
-0.148 |
| 199 |
255277_at |
AT4G04890
|
PDF2, protodermal factor 2 |
-0.154 |
-0.057 |
| 200 |
254214_at |
AT4G23640
|
ATKT3, KUP4, TRH1, TINY ROOT HAIR 1 |
-0.153 |
0.054 |
| 201 |
250398_at |
AT5G11000
|
unknown |
-0.153 |
-0.027 |
| 202 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.152 |
-0.126 |
| 203 |
256706_at |
AT3G30300
|
[O-fucosyltransferase family protein] |
-0.152 |
0.011 |
| 204 |
248413_at |
AT5G51600
|
ATMAP65-3, ARABIDOPSIS THALIANA MICROTUBULE-ASSOCIATED PROTEIN 65-3, MAP65-3, MICROTUBULE-ASSOCIATED PROTEIN 65-3, PLE, PLEIADE |
-0.151 |
0.103 |
| 205 |
267619_at |
AT2G26730
|
[Leucine-rich repeat protein kinase family protein] |
-0.148 |
-0.062 |
| 206 |
261874_at |
AT1G50640
|
ATERF3, ERF3, ethylene responsive element binding factor 3 |
-0.147 |
0.107 |
| 207 |
260733_at |
AT1G17640
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.147 |
-0.040 |
| 208 |
262419_at |
AT1G50380
|
[Prolyl oligopeptidase family protein] |
-0.147 |
0.023 |
| 209 |
261621_at |
AT1G01960
|
EDA10, embryo sac development arrest 10 |
-0.146 |
0.277 |
| 210 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
-0.146 |
-0.064 |
| 211 |
258480_at |
AT3G02640
|
unknown |
-0.144 |
0.025 |
| 212 |
259694_at |
AT1G63180
|
UGE3, UDP-D-glucose/UDP-D-galactose 4-epimerase 3 |
-0.144 |
-0.001 |
| 213 |
265767_at |
AT2G48110
|
MED33B, REF4, REDUCED EPIDERMAL FLUORESCENCE 4 |
-0.144 |
0.023 |
| 214 |
262297_at |
AT1G27600
|
I9H, IRREGULAR XYLEM 9 Homolog, IRX9-L, IRREGULAR XYLEM 9-LIKE |
-0.143 |
-0.088 |
| 215 |
252856_at |
AT4G39690
|
unknown |
-0.141 |
0.151 |
| 216 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.139 |
0.424 |
| 217 |
265219_at |
AT2G02050
|
[NADH-ubiquinone oxidoreductase B18 subunit, putative] |
-0.139 |
-0.092 |