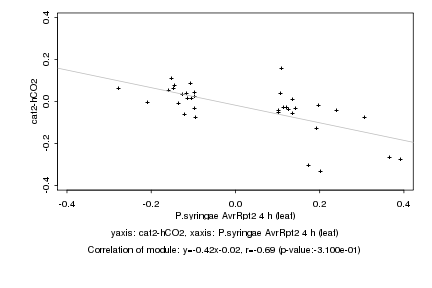
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
P.syringae AvrRpt2 4 h (leaf) |
Log2 signal ratio
cat2-hCO2 |
| 1 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
0.391 |
-0.275 |
| 2 |
262177_at |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
0.365 |
-0.263 |
| 3 |
262731_at |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
0.305 |
-0.073 |
| 4 |
254905_at |
AT4G11170
|
RMG1, Resistance Methylated Gene 1 |
0.239 |
-0.040 |
| 5 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
0.200 |
-0.332 |
| 6 |
254211_at |
AT4G23570
|
SGT1A |
0.196 |
-0.018 |
| 7 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
0.191 |
-0.127 |
| 8 |
246099_at |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
0.173 |
-0.304 |
| 9 |
250279_at |
AT5G13200
|
[GRAM domain family protein] |
0.141 |
-0.030 |
| 10 |
256797_at |
AT3G18600
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.135 |
0.011 |
| 11 |
260602_at |
AT1G55920
|
ATSERAT2;1, serine acetyltransferase 2;1, SAT1, SERINE ACETYLTRANSFERASE 1, SAT5, SERINE ACETYLTRANSFERASE 5, SERAT2;1, serine acetyltransferase 2;1 |
0.134 |
-0.057 |
| 12 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.125 |
-0.038 |
| 13 |
251538_at |
AT3G58660
|
[Ribosomal protein L1p/L10e family] |
0.120 |
-0.024 |
| 14 |
249719_at |
AT5G35735
|
[Auxin-responsive family protein] |
0.112 |
-0.028 |
| 15 |
262801_at |
AT1G21010
|
unknown |
0.109 |
0.162 |
| 16 |
251800_at |
AT3G55510
|
RBL, REBELOTE |
0.106 |
0.041 |
| 17 |
259439_at |
AT1G01480
|
ACS2, 1-amino-cyclopropane-1-carboxylate synthase 2, AT-ACC2 |
0.101 |
-0.041 |
| 18 |
252939_at |
AT4G39230
|
[NmrA-like negative transcriptional regulator family protein] |
0.101 |
-0.049 |
| 19 |
253743_at |
AT4G28940
|
[Phosphorylase superfamily protein] |
-0.280 |
0.066 |
| 20 |
256816_at |
AT3G21400
|
unknown |
-0.211 |
-0.004 |
| 21 |
259171_at |
AT3G03590
|
[SWIB/MDM2 domain superfamily protein] |
-0.161 |
0.057 |
| 22 |
252215_at |
AT3G50240
|
KICP-02 |
-0.153 |
0.112 |
| 23 |
246817_at |
AT5G27240
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.149 |
0.066 |
| 24 |
264978_at |
AT1G27120
|
[Galactosyltransferase family protein] |
-0.147 |
0.079 |
| 25 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.136 |
-0.007 |
| 26 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.127 |
0.038 |
| 27 |
264187_at |
AT1G54860
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.123 |
-0.058 |
| 28 |
249553_at |
AT5G38260
|
[Protein kinase superfamily protein] |
-0.117 |
0.043 |
| 29 |
249507_at |
AT5G38370
|
[FUNCTIONS IN: zinc ion binding] |
-0.114 |
0.015 |
| 30 |
262680_at |
AT1G75880
|
[SGNH hydrolase-type esterase superfamily protein] |
-0.107 |
0.087 |
| 31 |
265255_at |
AT2G28420
|
GLYI8, glyoxylase I 8 |
-0.106 |
0.019 |
| 32 |
266324_at |
AT2G46710
|
ROPGAP3, ROP guanosine triphosphatase (GTPase)-activating protein 3 |
-0.099 |
0.026 |
| 33 |
246015_at |
AT5G10700
|
[Peptidyl-tRNA hydrolase II (PTH2) family protein] |
-0.099 |
0.045 |
| 34 |
245959_at |
AT5G19640
|
[Major facilitator superfamily protein] |
-0.099 |
-0.031 |
| 35 |
246524_at |
AT5G15860
|
ATPCME, prenylcysteine methylesterase, ICME, Isoprenylcysteine methylesterase, PCME, prenylcysteine methylesterase |
-0.097 |
-0.072 |