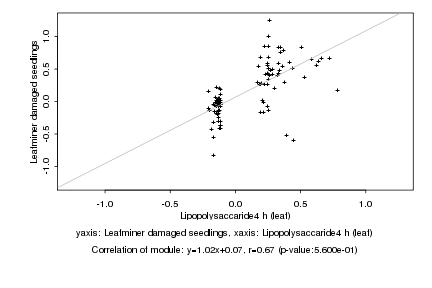
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Lipopolysaccaride4 h (leaf) |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
246366_at |
AT1G51850
|
[Leucine-rich repeat protein kinase family protein] |
0.778 |
0.173 |
| 2 |
252511_at |
AT3G46280
|
[protein kinase-related] |
0.717 |
0.678 |
| 3 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
0.660 |
0.677 |
| 4 |
264616_at |
AT2G17740
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.630 |
0.623 |
| 5 |
266967_at |
AT2G39530
|
[Uncharacterised protein family (UPF0497)] |
0.618 |
0.562 |
| 6 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
0.582 |
0.650 |
| 7 |
252045_at |
AT3G52450
|
AtPUB22, PUB22, plant U-box 22 |
0.528 |
0.373 |
| 8 |
254447_at |
AT4G20860
|
[FAD-binding Berberine family protein] |
0.499 |
0.837 |
| 9 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
0.438 |
-0.588 |
| 10 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.436 |
0.514 |
| 11 |
248932_at |
AT5G46050
|
AtNPF5.2, ATPTR3, ARABIDOPSIS THALIANA PEPTIDE TRANSPORTER 3, NPF5.2, NRT1/ PTR family 5.2, PTR3, peptide transporter 3 |
0.412 |
0.616 |
| 12 |
248657_at |
AT5G48570
|
ATFKBP65, FKBP65, ROF2 |
0.390 |
-0.516 |
| 13 |
266247_at |
AT2G27660
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.370 |
0.294 |
| 14 |
258209_at |
AT3G14060
|
unknown |
0.366 |
0.801 |
| 15 |
257623_at |
AT3G26210
|
CYP71B23, cytochrome P450, family 71, subfamily B, polypeptide 23 |
0.355 |
0.544 |
| 16 |
256576_at |
AT3G28210
|
PMZ, SAP12, STRESS-ASSOCIATED PROTEIN 12 |
0.343 |
0.839 |
| 17 |
256442_at |
AT3G10930
|
unknown |
0.340 |
0.757 |
| 18 |
251054_at |
AT5G01540
|
LecRK-VI.2, L-type lectin receptor kinase-VI.2, LECRKA4.1, lectin receptor kinase a4.1 |
0.337 |
0.487 |
| 19 |
247949_at |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
0.328 |
0.597 |
| 20 |
253173_at |
AT4G35110
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
0.324 |
0.837 |
| 21 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.323 |
0.444 |
| 22 |
264635_at |
AT1G65500
|
unknown |
0.316 |
0.405 |
| 23 |
249125_at |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.295 |
0.207 |
| 24 |
254453_at |
AT4G21120
|
AAT1, amino acid transporter 1, CAT1, CATIONIC AMINO ACID TRANSPORTER 1 |
0.284 |
0.419 |
| 25 |
252303_at |
AT3G49210
|
[O-acyltransferase (WSD1-like) family protein] |
0.282 |
0.507 |
| 26 |
251895_at |
AT3G54420
|
ATCHITIV, CHITINASE CLASS IV, ATEP3, homolog of carrot EP3-3 chitinase, CHIV, EP3, homolog of carrot EP3-3 chitinase |
0.268 |
0.484 |
| 27 |
248298_at |
AT5G53110
|
[RING/U-box superfamily protein] |
0.256 |
0.404 |
| 28 |
259879_at |
AT1G76650
|
CML38, calmodulin-like 38 |
0.256 |
1.263 |
| 29 |
245136_at |
AT2G45210
|
SAG201, senescence-associated gene 201, SAUR36, SMALL AUXIN UPREGULATED 36 |
0.253 |
0.354 |
| 30 |
247740_at |
AT5G58940
|
CRCK1, calmodulin-binding receptor-like cytoplasmic kinase 1 |
0.252 |
0.424 |
| 31 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.251 |
1.003 |
| 32 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
0.251 |
-0.132 |
| 33 |
264042_at |
AT2G03760
|
AtSOT1, AtSOT12, sulphotransferase 12, ATST1, ARABIDOPSIS THALIANA SULFOTRANSFERASE 1, AtSULT202A1, RAR047, SOT12, sulphotransferase 12, ST, ST1, SULFOTRANSFERASE 1, SULT202A1, sulfotransferase 202A1 |
0.251 |
0.690 |
| 34 |
252984_at |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
0.247 |
0.853 |
| 35 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.247 |
0.509 |
| 36 |
252334_at |
AT3G48850
|
MPT2, mitochondrial phosphate transporter 2, PHT3;2, phosphate transporter 3;2 |
0.244 |
0.586 |
| 37 |
256337_at |
AT1G72060
|
[serine-type endopeptidase inhibitors] |
0.243 |
0.559 |
| 38 |
256969_at |
AT3G21080
|
[ABC transporter-related] |
0.243 |
0.272 |
| 39 |
264774_at |
AT1G22890
|
unknown |
0.242 |
0.444 |
| 40 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
0.241 |
-0.071 |
| 41 |
248381_at |
AT5G51830
|
[pfkB-like carbohydrate kinase family protein] |
0.224 |
0.430 |
| 42 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
0.220 |
0.861 |
| 43 |
247610_at |
AT5G60630
|
unknown |
0.217 |
0.276 |
| 44 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.215 |
-0.007 |
| 45 |
253571_at |
AT4G31000
|
[Calmodulin-binding protein] |
0.211 |
-0.159 |
| 46 |
261727_at |
AT1G76090
|
SMT3, sterol methyltransferase 3 |
0.200 |
0.030 |
| 47 |
254410_at |
AT4G21410
|
CRK29, cysteine-rich RLK (RECEPTOR-like protein kinase) 29 |
0.195 |
0.289 |
| 48 |
246373_at |
AT1G51860
|
[Leucine-rich repeat protein kinase family protein] |
0.191 |
0.266 |
| 49 |
263314_at |
AT2G05760
|
[Xanthine/uracil permease family protein] |
0.187 |
-0.160 |
| 50 |
245317_at |
AT4G15610
|
[Uncharacterised protein family (UPF0497)] |
0.187 |
0.693 |
| 51 |
264767_at |
AT1G61380
|
SD1-29, S-domain-1 29 |
0.180 |
0.264 |
| 52 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
0.175 |
0.543 |
| 53 |
256627_at |
AT3G19970
|
[alpha/beta-Hydrolases superfamily protein] |
0.168 |
0.294 |
| 54 |
245445_at |
AT4G16750
|
[Integrase-type DNA-binding superfamily protein] |
-0.214 |
0.155 |
| 55 |
258232_at |
AT3G27750
|
EMB3123, EMBRYO DEFECTIVE 3123, THA8, Thylakoid Assembly 8 |
-0.212 |
-0.099 |
| 56 |
265828_at |
AT2G14520
|
unknown |
-0.205 |
-0.131 |
| 57 |
253097_at |
AT4G37320
|
CYP81D5, cytochrome P450, family 81, subfamily D, polypeptide 5 |
-0.184 |
-0.425 |
| 58 |
251509_at |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-0.174 |
-0.033 |
| 59 |
256237_at |
AT3G12610
|
DRT100, DNA-DAMAGE REPAIR/TOLERATION 100 |
-0.173 |
-0.829 |
| 60 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.170 |
-0.548 |
| 61 |
258033_at |
AT3G21250
|
ABCC8, ATP-binding cassette C8, ATMRP6, ARABIDOPSIS THALIANA MULTIDRUG RESISTANCE-ASSOCIATED PROTEIN 6, MRP6, multidrug resistance-associated protein 6 |
-0.170 |
-0.310 |
| 62 |
262181_at |
AT1G78060
|
[Glycosyl hydrolase family protein] |
-0.168 |
-0.149 |
| 63 |
247615_at |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
-0.162 |
-0.055 |
| 64 |
253356_at |
AT4G33390
|
unknown |
-0.160 |
-0.015 |
| 65 |
256105_at |
AT1G16910
|
LSH8, LIGHT SENSITIVE HYPOCOTYLS 8 |
-0.154 |
0.073 |
| 66 |
264972_at |
AT1G67370
|
ASY1, ASYNAPTIC 1, ATASY1 |
-0.151 |
-0.015 |
| 67 |
247671_at |
AT5G60210
|
RIP5, ROP interactive partner 5 |
-0.151 |
-0.178 |
| 68 |
250045_at |
AT5G17700
|
[MATE efflux family protein] |
-0.148 |
-0.179 |
| 69 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.148 |
0.227 |
| 70 |
248194_at |
AT5G54095
|
unknown |
-0.147 |
0.020 |
| 71 |
251485_at |
AT3G59550
|
ATRAD21.2, SISTER CHROMATID COHESION 1 PROTEIN 3, ATSYN3, SYN3 |
-0.146 |
-0.069 |
| 72 |
252698_at |
AT3G43670
|
[Copper amine oxidase family protein] |
-0.141 |
0.020 |
| 73 |
251050_at |
AT5G02440
|
unknown |
-0.141 |
-0.186 |
| 74 |
266915_at |
AT2G45870
|
[Bestrophin-like protein] |
-0.141 |
-0.060 |
| 75 |
265504_at |
AT2G15520
|
[similar to zinc finger protein, putative [Arabidopsis thaliana] (TAIR:AT2G15520.1)] |
-0.140 |
-0.020 |
| 76 |
264176_at |
AT1G02110
|
unknown |
-0.135 |
-0.304 |
| 77 |
261374_at |
AT1G52990
|
[thioredoxin family protein] |
-0.134 |
-0.025 |
| 78 |
245171_at |
AT2G47560
|
[RING/U-box superfamily protein] |
-0.134 |
-0.149 |
| 79 |
254789_at |
AT4G12880
|
AtENODL19, ENODL19, early nodulin-like protein 19 |
-0.134 |
-0.123 |
| 80 |
265647_at |
AT2G27410
|
unknown |
-0.133 |
-0.021 |
| 81 |
247681_at |
AT5G59640
|
[CACTA-like transposase family (En/Spm), has a 5.1e-159 P-value blast match to GB:AAD55677 putative transposase protein (CACTA-element) transposon=Shooter (Zea mays)] |
-0.133 |
-0.070 |
| 82 |
261804_at |
AT1G30530
|
UGT78D1, UDP-glucosyl transferase 78D1 |
-0.133 |
-0.235 |
| 83 |
246551_at |
AT5G15070
|
[Phosphoglycerate mutase-like family protein] |
-0.132 |
-0.180 |
| 84 |
246939_at |
AT5G25390
|
SHN3, shine3 |
-0.130 |
0.047 |
| 85 |
248725_at |
AT5G47980
|
[HXXXD-type acyl-transferase family protein] |
-0.130 |
0.005 |
| 86 |
265484_at |
AT2G15820
|
OTP51, ORGANELLE TRANSCRIPT PROCESSING 51 |
-0.130 |
-0.306 |
| 87 |
245689_at |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
-0.128 |
-0.043 |
| 88 |
250802_at |
AT5G04970
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.127 |
0.022 |
| 89 |
258355_at |
AT3G14330
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.127 |
-0.408 |
| 90 |
250719_at |
AT5G06250
|
DPA4, DEVELOPMENT-RELATED PcG TARGET IN THE APEX 4 |
-0.127 |
-0.045 |
| 91 |
251271_at |
AT3G62050
|
[Putative endonuclease or glycosyl hydrolase] |
-0.125 |
-0.136 |
| 92 |
251554_at |
AT3G58670
|
unknown |
-0.123 |
0.060 |
| 93 |
266326_at |
AT2G46650
|
ATCB5-C, ARABIDOPSIS CYTOCHROME B5 ISOFORM C, B5 #1, CB5-C, cytochrome B5 isoform C, CYTB5-C |
-0.123 |
0.204 |
| 94 |
265601_at |
AT2G14390
|
unknown |
-0.122 |
-0.036 |
| 95 |
250633_at |
AT5G07460
|
ATMSRA2, ARABIDOPSIS THALIANA METHIONINE SULFOXIDE REDUCTASE 2, PMSR2, peptidemethionine sulfoxide reductase 2 |
-0.120 |
0.197 |
| 96 |
251395_at |
AT2G45470
|
AGP8, ARABINOGALACTAN PROTEIN 8, FLA8, FASCICLIN-like arabinogalactan protein 8 |
-0.120 |
-0.363 |
| 97 |
263793_at |
AT2G24630
|
ATCSLC08, ATCSLC8, CELLULOSE-SYNTHASE LIKE C8, CSLC08 |
-0.120 |
-0.406 |
| 98 |
265065_at |
AT1G03980
|
ATPCS2, phytochelatin synthase 2, PCS2, phytochelatin synthase 2 |
-0.118 |
0.027 |
| 99 |
254011_at |
AT4G26370
|
[antitermination NusB domain-containing protein] |
-0.118 |
0.111 |
| 100 |
246047_at |
AT5G28870
|
[pseudogene] |
-0.117 |
-0.074 |
| 101 |
251133_at |
AT5G01240
|
LAX1, like AUXIN RESISTANT 1 |
-0.117 |
-0.354 |
| 102 |
252420_at |
AT3G47530
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.116 |
-0.005 |
| 103 |
252797_at |
AT3G42300
|
unknown |
-0.116 |
-0.040 |
| 104 |
265121_at |
AT1G62560
|
FMO GS-OX3, flavin-monooxygenase glucosinolate S-oxygenase 3 |
-0.115 |
-0.299 |
| 105 |
259291_at |
AT3G11550
|
CASP2, Casparian strip membrane domain protein 2 |
-0.115 |
-0.041 |