|
probeID |
AGICode |
Annotation |
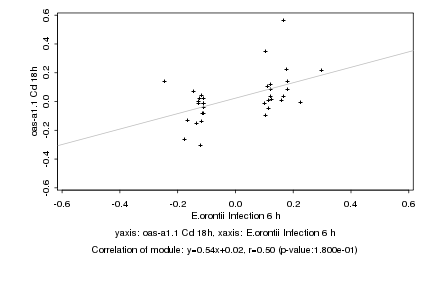
Log2 signal ratio
E.orontii Infection 6 h |
Log2 signal ratio
oas-a1.1 Cd 18h |
| 1 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.297 |
0.221 |
| 2 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.225 |
0.000 |
| 3 |
260472_at |
AT1G10990
|
unknown |
0.180 |
0.086 |
| 4 |
266017_at |
AT2G18690
|
unknown |
0.177 |
0.146 |
| 5 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
0.174 |
0.227 |
| 6 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
0.166 |
0.570 |
| 7 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.163 |
0.035 |
| 8 |
252989_at |
AT4G38420
|
sks9, SKU5 similar 9 |
0.157 |
0.008 |
| 9 |
266951_at |
AT2G18940
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.122 |
0.021 |
| 10 |
246574_at |
AT1G31670
|
[Copper amine oxidase family protein] |
0.120 |
0.123 |
| 11 |
249546_at |
AT5G38150
|
PMI15, plastid movement impaired 15 |
0.119 |
0.036 |
| 12 |
265713_at |
AT2G03530
|
ATUPS2, ARABIDOPSIS THALIANA UREIDE PERMEASE 2, UPS2, ureide permease 2 |
0.118 |
0.088 |
| 13 |
251750_at |
AT3G55710
|
[UDP-Glycosyltransferase superfamily protein] |
0.112 |
-0.046 |
| 14 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.112 |
0.008 |
| 15 |
262120_at |
AT1G02950
|
ATGSTF4, glutathione S-transferase F4, GST31, GLUTATHIONE S-TRANSFERASE 31, GSTF4, glutathione S-transferase F4 |
0.108 |
0.108 |
| 16 |
260799_at |
AT1G78270
|
AtUGT85A4, UDP-glucosyl transferase 85A4, UGT85A4, UDP-glucosyl transferase 85A4 |
0.103 |
-0.096 |
| 17 |
254241_at |
AT4G23190
|
AT-RLK3, RECEPTOR LIKE PROTEIN KINASE 3, CRK11, cysteine-rich RLK (RECEPTOR-like protein kinase) 11 |
0.102 |
0.352 |
| 18 |
261901_at |
AT1G80920
|
AtJ8, AtToc12, translocon at the outer envelope membrane of chloroplasts 12, DJC22, DNA J protein C22, J8, Toc12, translocon at the outer envelope membrane of chloroplasts 12 |
0.099 |
-0.010 |
| 19 |
247240_at |
AT5G64660
|
ATCMPG2, CMPG2, CYS, MET, PRO, and GLY protein 2 |
-0.249 |
0.145 |
| 20 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
-0.179 |
-0.263 |
| 21 |
254371_at |
AT4G21760
|
BGLU47, beta-glucosidase 47 |
-0.169 |
-0.129 |
| 22 |
248392_at |
AT5G52050
|
[MATE efflux family protein] |
-0.146 |
0.072 |
| 23 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.137 |
-0.151 |
| 24 |
263696_at |
AT1G31230
|
AK-HSDH, ASPARTATE KINASE-HOMOSERINE DEHYDROGENASE, AK-HSDH I, aspartate kinase-homoserine dehydrogenase i |
-0.129 |
-0.008 |
| 25 |
261913_at |
AT1G65860
|
FMO GS-OX1, flavin-monooxygenase glucosinolate S-oxygenase 1 |
-0.129 |
0.006 |
| 26 |
253274_at |
AT4G34200
|
EDA9, embryo sac development arrest 9, PGDH1, phosphoglycerate dehydrogenase 1 |
-0.126 |
0.023 |
| 27 |
251524_at |
AT3G58990
|
IPMI1, isopropylmalate isomerase 1 |
-0.123 |
-0.299 |
| 28 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
-0.120 |
0.048 |
| 29 |
260902_at |
AT1G21440
|
[Phosphoenolpyruvate carboxylase family protein] |
-0.119 |
-0.135 |
| 30 |
250859_at |
AT5G04660
|
CYP77A4, cytochrome P450, family 77, subfamily A, polypeptide 4 |
-0.117 |
-0.077 |
| 31 |
257213_at |
AT3G15020
|
mMDH2, mitochondrial malate dehydrogenase 2 |
-0.114 |
-0.007 |
| 32 |
252870_at |
AT4G39940
|
AKN2, APS-kinase 2, APK2, ADENOSINE-5'-PHOSPHOSULFATE (APS) KINASE 2 |
-0.113 |
-0.078 |
| 33 |
251592_at |
AT3G57670
|
NTT, NO TRANSMITTING TRACT, WIP2, WIP domain protein 2 |
-0.112 |
0.025 |
| 34 |
256931_at |
AT3G22490
|
[Seed maturation protein] |
-0.112 |
-0.011 |
| 35 |
247393_at |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
-0.111 |
-0.036 |