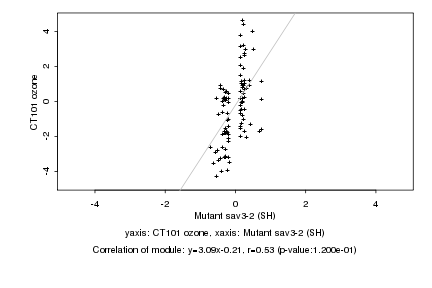
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant sav3-2 (SH) |
Log2 signal ratio
CT101 ozone |
| 1 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
0.741 |
-1.580 |
| 2 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.740 |
1.188 |
| 3 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
0.720 |
0.142 |
| 4 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.672 |
-1.676 |
| 5 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
0.495 |
2.992 |
| 6 |
246293_at |
AT3G56710
|
SIB1, sigma factor binding protein 1 |
0.485 |
4.006 |
| 7 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
0.425 |
-1.299 |
| 8 |
259352_at |
AT3G05170
|
[Phosphoglycerate mutase family protein] |
0.394 |
1.247 |
| 9 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
0.383 |
0.967 |
| 10 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
0.312 |
0.748 |
| 11 |
261221_at |
AT1G19960
|
unknown |
0.304 |
-2.032 |
| 12 |
249893_at |
AT5G22555
|
unknown |
0.261 |
3.014 |
| 13 |
267076_at |
AT2G41090
|
[Calcium-binding EF-hand family protein] |
0.255 |
1.236 |
| 14 |
249094_at |
AT5G43890
|
SUPER1, SUPPRESSOR OF ER 1, YUC5, YUCCA5 |
0.249 |
0.736 |
| 15 |
249780_at |
AT5G24240
|
[Phosphatidylinositol 3- and 4-kinase ] |
0.248 |
2.670 |
| 16 |
245074_at |
AT2G23200
|
[Protein kinase superfamily protein] |
0.245 |
0.267 |
| 17 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
0.245 |
-1.668 |
| 18 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.242 |
2.788 |
| 19 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.238 |
1.037 |
| 20 |
256100_at |
AT1G13750
|
[Purple acid phosphatases superfamily protein] |
0.238 |
-0.421 |
| 21 |
265611_at |
AT2G25510
|
unknown |
0.223 |
-0.998 |
| 22 |
259129_at |
AT3G02150
|
PTF1, plastid transcription factor 1, TCP13, TEOSINTE BRANCHED1, CYCLOIDEA AND PCF TRANSCRIPTION FACTOR 13, TFPD |
0.216 |
1.907 |
| 23 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
0.215 |
4.413 |
| 24 |
260419_at |
AT1G69730
|
[Wall-associated kinase family protein] |
0.212 |
0.510 |
| 25 |
255148_at |
AT4G08470
|
MAPKKK10, MEKK3, MAPK/ERK kinase kinase 3 |
0.209 |
0.958 |
| 26 |
263800_at |
AT2G24600
|
[Ankyrin repeat family protein] |
0.202 |
3.241 |
| 27 |
257646_at |
AT3G25740
|
MAP1B, methionine aminopeptidase 1C, MAP1C, METHIONINE AMINOPEPTIDASE 1C |
0.195 |
-0.763 |
| 28 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.192 |
0.190 |
| 29 |
252170_at |
AT3G50480
|
HR4, homolog of RPW8 4 |
0.184 |
4.689 |
| 30 |
254112_at |
AT4G24970
|
[Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase family protein] |
0.181 |
0.014 |
| 31 |
253696_at |
AT4G29740
|
ATCKX4, CKX4, cytokinin oxidase 4 |
0.181 |
0.952 |
| 32 |
262508_at |
AT1G11300
|
[protein serine/threonine kinases] |
0.177 |
0.840 |
| 33 |
248270_at |
AT5G53450
|
ORG1, OBP3-responsive gene 1 |
0.177 |
-0.041 |
| 34 |
252427_at |
AT3G47640
|
PYE, POPEYE |
0.161 |
-0.427 |
| 35 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
0.158 |
1.033 |
| 36 |
245227_s_at |
AT1G08410
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.153 |
0.000 |
| 37 |
255503_at |
AT4G02420
|
LecRK-IV.4, L-type lectin receptor kinase IV.4 |
0.149 |
-1.246 |
| 38 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
0.146 |
1.200 |
| 39 |
254289_at |
AT4G22980
|
unknown |
0.138 |
3.813 |
| 40 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.134 |
-1.966 |
| 41 |
253993_at |
AT4G26070
|
ATMEK1, MEK1, MAP kinase/ ERK kinase 1, MKK1, MITOGEN ACTIVATED PROTEIN KINASE KINASE 1, NMAPKK |
0.133 |
1.544 |
| 42 |
267084_at |
AT2G41180
|
SIB2, sigma factor binding protein 2 |
0.133 |
0.618 |
| 43 |
264901_at |
AT1G23090
|
AST91, sulfate transporter 91, SULTR3;3 |
0.127 |
3.193 |
| 44 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.123 |
2.094 |
| 45 |
251569_at |
AT3G58290
|
[TRAF-like superfamily protein] |
0.123 |
0.225 |
| 46 |
249032_at |
AT5G44910
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.123 |
2.562 |
| 47 |
254639_at |
AT4G18750
|
DOT4, DEFECTIVELY ORGANIZED TRIBUTARIES 4 |
0.123 |
-0.197 |
| 48 |
254758_at |
AT4G13260
|
AtYUC2, YUC2, YUCCA2 |
0.121 |
-1.384 |
| 49 |
262286_at |
AT1G68585
|
unknown |
0.120 |
-0.630 |
| 50 |
255852_at |
AT1G66970
|
GDPDL1, Glycerophosphodiester phosphodiesterase (GDPD) like 1, SVL2, SHV3-like 2 |
0.119 |
-0.488 |
| 51 |
261598_at |
AT1G49750
|
[Leucine-rich repeat (LRR) family protein] |
0.116 |
-1.495 |
| 52 |
251792_at |
AT3G55550
|
LecRK-S.4, L-type lectin receptor kinase S.4 |
0.116 |
0.225 |
| 53 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.735 |
-2.583 |
| 54 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.636 |
-3.494 |
| 55 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.596 |
-2.891 |
| 56 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.564 |
-4.283 |
| 57 |
263890_at |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-0.554 |
0.225 |
| 58 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.515 |
-2.788 |
| 59 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
-0.503 |
-0.695 |
| 60 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.498 |
-3.341 |
| 61 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.451 |
-3.209 |
| 62 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
-0.445 |
0.946 |
| 63 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.439 |
0.778 |
| 64 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.412 |
-3.989 |
| 65 |
264100_at |
AT1G78970
|
ATLUP1, ARABIDOPSIS THALIANA LUPEOL SYNTHASE 1, LUP1, lupeol synthase 1 |
-0.397 |
-1.837 |
| 66 |
247474_at |
AT5G62280
|
unknown |
-0.390 |
-0.573 |
| 67 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.382 |
0.046 |
| 68 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.378 |
-2.579 |
| 69 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.366 |
0.740 |
| 70 |
251178_at |
AT3G63440
|
ATCKX6, CYTOKININ OXIDASE 6, ATCKX7, CKX6, cytokinin oxidase/dehydrogenase 6 |
-0.364 |
-0.220 |
| 71 |
245076_at |
AT2G23170
|
GH3.3 |
-0.351 |
0.206 |
| 72 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
-0.337 |
-3.182 |
| 73 |
248801_at |
AT5G47370
|
HAT2 |
-0.335 |
-1.705 |
| 74 |
252765_at |
AT3G42800
|
unknown |
-0.330 |
-1.819 |
| 75 |
254685_at |
AT4G13790
|
SAUR25, SMALL AUXIN UPREGULATED RNA 25 |
-0.317 |
0.225 |
| 76 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-0.316 |
0.231 |
| 77 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.301 |
-3.174 |
| 78 |
252204_at |
AT3G50340
|
unknown |
-0.298 |
0.075 |
| 79 |
256464_at |
AT1G32560
|
AtLEA4-1, Late Embryogenesis Abundant 4-1, LEA4-1, Late Embryogenesis Abundant 4-1 |
-0.297 |
0.530 |
| 80 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.297 |
-1.783 |
| 81 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.296 |
-2.712 |
| 82 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.296 |
-1.823 |
| 83 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.294 |
0.068 |
| 84 |
250868_at |
AT5G03860
|
MLS, malate synthase |
-0.294 |
-1.542 |
| 85 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.292 |
-3.119 |
| 86 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
-0.283 |
0.225 |
| 87 |
264598_at |
AT1G04610
|
YUC3, YUCCA 3 |
-0.278 |
0.602 |
| 88 |
248104_at |
AT5G55250
|
AtIAMT1, IAMT1, IAA carboxylmethyltransferase 1 |
-0.259 |
-1.686 |
| 89 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.255 |
-1.702 |
| 90 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.246 |
-3.894 |
| 91 |
247159_at |
AT5G65800
|
ACS5, ACC synthase 5, ATACS5, CIN5, CYTOKININ-INSENSITIVE 5, ETO2, ETHYLENE OVERPRODUCER 2 |
-0.236 |
-1.078 |
| 92 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.236 |
-1.752 |
| 93 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.228 |
-0.632 |
| 94 |
250396_at |
AT5G10970
|
[C2H2 and C2HC zinc fingers superfamily protein] |
-0.224 |
-2.104 |
| 95 |
255028_at |
AT4G09890
|
unknown |
-0.219 |
-2.284 |
| 96 |
257642_at |
AT3G25710
|
ATAIG1, BHLH32, basic helix-loop-helix 32, TMO5, TARGET OF MONOPTEROS 5 |
-0.218 |
-1.881 |
| 97 |
250648_at |
AT5G06760
|
AtLEA4-5, Late Embryogenesis Abundant 4-5, LEA4-5, Late Embryogenesis Abundant 4-5 |
-0.209 |
-1.419 |
| 98 |
260716_at |
AT1G48130
|
ATPER1, 1-cysteine peroxiredoxin 1, PER1, 1-cysteine peroxiredoxin 1 |
-0.202 |
0.225 |
| 99 |
264016_at |
AT2G21220
|
SAUR12, SMALL AUXIN UPREGULATED RNA 12 |
-0.202 |
-0.986 |
| 100 |
258907_at |
AT3G06370
|
ATNHX4, NHX4, sodium hydrogen exchanger 4 |
-0.201 |
-0.027 |
| 101 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.201 |
-3.188 |
| 102 |
262373_at |
AT1G73120
|
unknown |
-0.199 |
0.490 |
| 103 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.194 |
-3.475 |