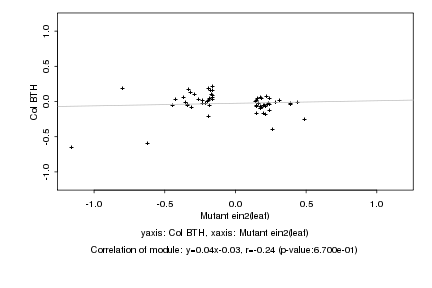
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant ein2(leaf) |
Log2 signal ratio
Col BTH |
| 1 |
256021_at |
AT1G58270
|
ZW9 |
0.483 |
-0.248 |
| 2 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.437 |
-0.012 |
| 3 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.387 |
-0.015 |
| 4 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.386 |
-0.031 |
| 5 |
245523_at |
AT4G15910
|
ATDI21, drought-induced 21, DI21, drought-induced 21 |
0.306 |
0.018 |
| 6 |
247467_at |
AT5G62130
|
[Per1-like family protein] |
0.280 |
-0.011 |
| 7 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
0.260 |
-0.395 |
| 8 |
266293_at |
AT2G29360
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.240 |
-0.039 |
| 9 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
0.238 |
-0.121 |
| 10 |
263544_at |
AT2G21590
|
APL4 |
0.234 |
0.046 |
| 11 |
263709_at |
AT1G09310
|
unknown |
0.227 |
-0.028 |
| 12 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.224 |
-0.034 |
| 13 |
249033_at |
AT5G44920
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
0.213 |
0.076 |
| 14 |
256509_at |
AT1G75300
|
[NmrA-like negative transcriptional regulator family protein] |
0.209 |
-0.178 |
| 15 |
266485_at |
AT2G47630
|
[alpha/beta-Hydrolases superfamily protein] |
0.208 |
-0.069 |
| 16 |
246908_at |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
0.199 |
-0.048 |
| 17 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.196 |
-0.053 |
| 18 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.195 |
-0.165 |
| 19 |
266225_at |
AT2G28900
|
ATOEP16-1, outer plastid envelope protein 16-1, ATOEP16-L, OUTER PLASTID ENVELOPE PROTEIN 16-L, OEP16, outer envelope protein 16, OEP16-1, outer plastid envelope protein 16-1 |
0.183 |
-0.076 |
| 20 |
266635_at |
AT2G35470
|
unknown |
0.183 |
0.046 |
| 21 |
250597_at |
AT5G07680
|
ANAC079, Arabidopsis NAC domain containing protein 79, ANAC080, NAC domain containing protein 80, ATNAC4, NAC080, NAC domain containing protein 80, NAC4 |
0.176 |
0.059 |
| 22 |
254225_at |
AT4G23670
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.175 |
-0.093 |
| 23 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
0.174 |
-0.094 |
| 24 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.174 |
-0.057 |
| 25 |
260238_at |
AT1G74520
|
ATHVA22A, HVA22 homologue A, HVA22A, HVA22 homologue A |
0.158 |
-0.027 |
| 26 |
255059_at |
AT4G09420
|
[Disease resistance protein (TIR-NBS class)] |
0.150 |
0.047 |
| 27 |
263513_at |
AT2G12400
|
unknown |
0.146 |
-0.063 |
| 28 |
254332_at |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
0.145 |
-0.156 |
| 29 |
261065_at |
AT1G07500
|
SMR5, SIAMESE-RELATED 5 |
0.144 |
0.015 |
| 30 |
247216_at |
AT5G64860
|
DPE1, disproportionating enzyme |
0.143 |
-0.051 |
| 31 |
262827_at |
AT1G13100
|
CYP71B29, cytochrome P450, family 71, subfamily B, polypeptide 29 |
0.141 |
0.006 |
| 32 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-1.165 |
-0.650 |
| 33 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
-0.803 |
0.189 |
| 34 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.623 |
-0.591 |
| 35 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
-0.446 |
-0.044 |
| 36 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.428 |
0.032 |
| 37 |
249997_at |
AT5G18620
|
CHR17, chromatin remodeling factor17 |
-0.373 |
0.067 |
| 38 |
246935_at |
AT5G25350
|
EBF2, EIN3-binding F box protein 2 |
-0.354 |
-0.012 |
| 39 |
258925_at |
AT3G10420
|
SPD1, SEEDLING PLASTID DEVELOPMENT 1 |
-0.344 |
-0.033 |
| 40 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.341 |
-0.053 |
| 41 |
261242_at |
AT1G32960
|
ATSBT3.3, SBT3.3 |
-0.337 |
0.173 |
| 42 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.319 |
0.135 |
| 43 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.315 |
-0.071 |
| 44 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.296 |
0.102 |
| 45 |
260140_at |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
-0.262 |
0.033 |
| 46 |
254574_at |
AT4G19430
|
unknown |
-0.238 |
-0.018 |
| 47 |
257178_at |
AT3G13070
|
[CBS domain-containing protein / transporter associated domain-containing protein] |
-0.236 |
0.027 |
| 48 |
264698_at |
AT1G70200
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.218 |
-0.021 |
| 49 |
261052_at |
AT1G01440
|
unknown |
-0.199 |
0.003 |
| 50 |
267044_at |
AT2G34357
|
[ARM repeat superfamily protein] |
-0.197 |
0.028 |
| 51 |
266752_at |
AT2G47000
|
ABCB4, ATP-binding cassette B4, AtABCB4, ATPGP4, ARABIDOPSIS P-GLYCOPROTEIN 4, MDR4, MULTIDRUG RESISTANCE 4, PGP4, P-GLYCOPROTEIN 4 |
-0.195 |
0.194 |
| 52 |
254120_at |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
-0.192 |
-0.204 |
| 53 |
258884_at |
AT3G10050
|
OMR1, L-O-methylthreonine resistant 1 |
-0.190 |
-0.051 |
| 54 |
259718_at |
AT1G61040
|
VIP5, vernalization independence 5 |
-0.184 |
0.043 |
| 55 |
264851_at |
AT2G17290
|
ATCDPK3, ARABIDOPSIS THALIANA CALMODULIN-DOMAIN PROTEIN KINASE 3, ATCPK6, ARABIDOPSIS THALIANA CALCIUM-DEPENDENT PROTEIN KINASE 6, CPK6, calcium dependent protein kinase 6 |
-0.183 |
0.161 |
| 56 |
246529_at |
AT5G15730
|
AtCRLK2, CRLK2, calcium/calmodulin-regulated receptor-like kinase 2 |
-0.172 |
0.106 |
| 57 |
247500_at |
AT5G61910
|
[DCD (Development and Cell Death) domain protein] |
-0.169 |
0.062 |
| 58 |
265471_at |
AT2G37130
|
[Peroxidase superfamily protein] |
-0.167 |
0.030 |
| 59 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.165 |
0.227 |
| 60 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.164 |
0.169 |
| 61 |
259899_at |
AT1G71210
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.163 |
0.094 |