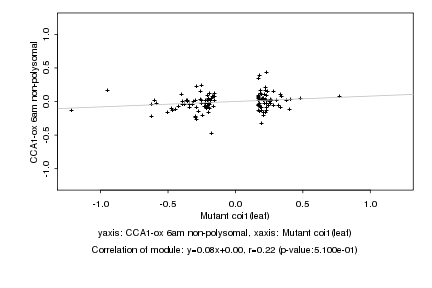
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant coi1(leaf) |
Log2 signal ratio
CCA1-ox 6am non-polysomal |
| 1 |
262128_at |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
0.768 |
0.084 |
| 2 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.476 |
0.049 |
| 3 |
252314_at |
AT3G49400
|
[Transducin/WD40 repeat-like superfamily protein] |
0.402 |
0.038 |
| 4 |
245162_at |
AT2G33240
|
ATXID, MYOSIN XI D, XID, myosin XI D |
0.397 |
-0.110 |
| 5 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.374 |
0.028 |
| 6 |
257129_at |
AT3G20100
|
CYP705A19, cytochrome P450, family 705, subfamily A, polypeptide 19 |
0.337 |
0.075 |
| 7 |
264909_at |
AT2G17300
|
unknown |
0.334 |
0.109 |
| 8 |
251026_at |
AT5G02200
|
FHL, far-red-elongated hypocotyl1-like |
0.330 |
-0.076 |
| 9 |
253943_at |
AT4G27030
|
FAD4, FATTY ACID DESATURASE 4, FADA, fatty acid desaturase A |
0.315 |
-0.057 |
| 10 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.301 |
0.027 |
| 11 |
249774_at |
AT5G24150
|
SQE5, SQUALENE MONOOXYGENASE 5, SQP1 |
0.280 |
-0.053 |
| 12 |
258055_at |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
0.276 |
0.150 |
| 13 |
258932_at |
AT3G10150
|
ATPAP16, PAP16, purple acid phosphatase 16 |
0.263 |
0.017 |
| 14 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
0.255 |
0.031 |
| 15 |
264078_at |
AT2G28470
|
BGAL8, beta-galactosidase 8 |
0.253 |
-0.025 |
| 16 |
255433_at |
AT4G03210
|
XTH9, xyloglucan endotransglucosylase/hydrolase 9 |
0.252 |
0.005 |
| 17 |
263442_at |
AT2G28605
|
[Photosystem II reaction center PsbP family protein] |
0.244 |
-0.046 |
| 18 |
255779_at |
AT1G18650
|
PDCB3, plasmodesmata callose-binding protein 3 |
0.232 |
0.155 |
| 19 |
251586_at |
AT3G58070
|
GIS, GLABROUS INFLORESCENCE STEMS |
0.230 |
0.435 |
| 20 |
267569_at |
AT2G30790
|
PSBP-2, photosystem II subunit P-2 |
0.227 |
-0.081 |
| 21 |
249410_at |
AT5G40380
|
CRK42, cysteine-rich RLK (RECEPTOR-like protein kinase) 42 |
0.226 |
-0.122 |
| 22 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
0.225 |
0.092 |
| 23 |
265128_at |
AT1G30860
|
[RING/U-box superfamily protein] |
0.225 |
-0.028 |
| 24 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.224 |
0.016 |
| 25 |
252366_at |
AT3G48420
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.224 |
0.029 |
| 26 |
258976_at |
AT3G01980
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.222 |
0.171 |
| 27 |
245088_at |
AT2G39850
|
[Subtilisin-like serine endopeptidase family protein] |
0.222 |
0.213 |
| 28 |
260481_at |
AT1G10960
|
ATFD1, ferredoxin 1, FD1, ferredoxin 1 |
0.220 |
0.035 |
| 29 |
261222_at |
AT1G20120
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.216 |
-0.161 |
| 30 |
261927_at |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
0.215 |
0.119 |
| 31 |
264751_at |
AT1G23020
|
ATFRO3, FERRIC REDUCTION OXIDASE 3, FRO3, ferric reduction oxidase 3 |
0.212 |
-0.026 |
| 32 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.206 |
0.023 |
| 33 |
262133_at |
AT1G78000
|
SEL1, SELENATE RESISTANT 1, SULTR1;2, sulfate transporter 1;2 |
0.205 |
-0.199 |
| 34 |
256655_at |
AT3G18890
|
AtTic62, translocon at the inner envelope membrane of chloroplasts 62, Tic62, translocon at the inner envelope membrane of chloroplasts 62 |
0.204 |
-0.158 |
| 35 |
259308_at |
AT3G05180
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.203 |
0.027 |
| 36 |
261488_at |
AT1G14345
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.203 |
0.032 |
| 37 |
263674_at |
AT2G04790
|
unknown |
0.202 |
0.057 |
| 38 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.202 |
0.048 |
| 39 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.195 |
0.053 |
| 40 |
254250_at |
AT4G23290
|
CRK21, cysteine-rich RLK (RECEPTOR-like protein kinase) 21 |
0.193 |
-0.321 |
| 41 |
246159_at |
AT5G20935
|
CRR42, CHLORORESPIRATORY REDUCTION 42 |
0.192 |
-0.081 |
| 42 |
260364_at |
AT1G70560
|
CKRC1, cytokinin induced root curling 1, SAV3, SHADE AVOIDANCE 3, TAA1, tryptophan aminotransferase of Arabidopsis 1, WEI8, WEAK ETHYLENE INSENSITIVE 8 |
0.190 |
0.032 |
| 43 |
248956_at |
AT5G45610
|
SUV2, SENSITIVE TO UV 2 |
0.187 |
0.111 |
| 44 |
250933_at |
AT5G03170
|
ATFLA11, ARABIDOPSIS FASCICLIN-LIKE ARABINOGALACTAN-PROTEIN 11, FLA11, FASCICLIN-like arabinogalactan-protein 11 |
0.187 |
-0.122 |
| 45 |
256983_at |
AT3G13470
|
Cpn60beta2, chaperonin-60beta2 |
0.185 |
0.132 |
| 46 |
255453_at |
AT4G02900
|
[ERD (early-responsive to dehydration stress) family protein] |
0.182 |
-0.082 |
| 47 |
257062_at |
AT3G18290
|
BTS, BRUTUS, EMB2454, embryo defective 2454 |
0.179 |
0.178 |
| 48 |
256386_at |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
0.178 |
-0.055 |
| 49 |
263038_at |
AT1G23270
|
unknown |
0.175 |
-0.137 |
| 50 |
250926_at |
AT5G03555
|
AtNCS1, NCS1, nucleobase cation symporter 1 |
0.174 |
0.040 |
| 51 |
249070_at |
AT5G44030
|
CESA4, cellulose synthase A4, IRX5, IRREGULAR XYLEM 5, NWS2 |
0.174 |
-0.074 |
| 52 |
247483_at |
AT5G62420
|
[NAD(P)-linked oxidoreductase superfamily protein] |
0.173 |
-0.022 |
| 53 |
262368_at |
AT1G73060
|
LPA3, Low PSII Accumulation 3 |
0.172 |
0.393 |
| 54 |
248651_at |
AT5G49260
|
unknown |
0.170 |
-0.038 |
| 55 |
267169_at |
AT2G37540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.170 |
0.022 |
| 56 |
251161_at |
AT3G63290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.169 |
-0.046 |
| 57 |
264340_at |
AT1G70280
|
[NHL domain-containing protein] |
0.169 |
0.098 |
| 58 |
250075_at |
AT5G17670
|
[alpha/beta-Hydrolases superfamily protein] |
0.168 |
0.050 |
| 59 |
248843_at |
AT5G46880
|
HB-7, homeobox-7, HDG5, HOMEODOMAIN GLABROUS 5 |
0.168 |
0.089 |
| 60 |
259081_at |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
0.166 |
-0.123 |
| 61 |
259203_at |
AT3G09130
|
unknown |
0.165 |
0.349 |
| 62 |
259742_at |
AT1G71120
|
GLIP6, GDSL-motif lipase/hydrolase 6 |
0.165 |
0.073 |
| 63 |
253518_at |
AT4G31400
|
AtCTF7, CTF7, CHROMOSOME TRANSMISSION FIDELITY 7, ECO1 |
0.164 |
-0.052 |
| 64 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-1.221 |
-0.128 |
| 65 |
266270_at |
AT2G29470
|
ATGSTU3, glutathione S-transferase tau 3, GST21, GLUTATHIONE S-TRANSFERASE 21, GSTU3, glutathione S-transferase tau 3 |
-0.950 |
0.179 |
| 66 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
-0.627 |
-0.038 |
| 67 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
-0.623 |
-0.209 |
| 68 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
-0.603 |
0.022 |
| 69 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-0.587 |
-0.015 |
| 70 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
-0.506 |
-0.156 |
| 71 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.477 |
-0.095 |
| 72 |
252200_at |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
-0.470 |
-0.128 |
| 73 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
-0.445 |
-0.116 |
| 74 |
266267_at |
AT2G29460
|
ATGSTU4, glutathione S-transferase tau 4, GST22, GLUTATHIONE S-TRANSFERASE 22, GSTU4, glutathione S-transferase tau 4 |
-0.424 |
-0.064 |
| 75 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.405 |
0.111 |
| 76 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
-0.398 |
0.009 |
| 77 |
253458_at |
AT4G32070
|
Phox4, Phox4 |
-0.396 |
-0.039 |
| 78 |
267026_at |
AT2G38340
|
DREB19, dehydration response element-binding protein 19 |
-0.382 |
-0.032 |
| 79 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
-0.370 |
0.027 |
| 80 |
260783_at |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-0.361 |
0.025 |
| 81 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.358 |
0.004 |
| 82 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
-0.346 |
-0.040 |
| 83 |
252114_at |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.343 |
-0.076 |
| 84 |
267518_at |
AT2G30500
|
NET4B, Networked 4B |
-0.321 |
-0.035 |
| 85 |
259730_at |
AT1G77660
|
[Histone H3 K4-specific methyltransferase SET7/9 family protein] |
-0.313 |
0.004 |
| 86 |
260425_at |
AT1G72440
|
EDA25, embryo sac development arrest 25, SWA2, SLOW WALKER2 |
-0.301 |
0.018 |
| 87 |
245041_at |
AT2G26530
|
AR781 |
-0.300 |
-0.226 |
| 88 |
260205_at |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
-0.299 |
-0.213 |
| 89 |
255733_at |
AT1G25400
|
unknown |
-0.295 |
-0.266 |
| 90 |
248713_at |
AT5G48180
|
AtNSP5, NSP5, nitrile specifier protein 5 |
-0.293 |
-0.084 |
| 91 |
259364_at |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-0.290 |
0.235 |
| 92 |
264339_at |
AT1G70290
|
ATTPS8, ATTPSC, TPS8, trehalose-6-phosphatase synthase S8 |
-0.275 |
-0.140 |
| 93 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
-0.265 |
0.154 |
| 94 |
258203_at |
AT3G13950
|
unknown |
-0.260 |
0.033 |
| 95 |
250875_at |
AT5G04020
|
[calmodulin binding] |
-0.258 |
0.017 |
| 96 |
265452_at |
AT2G46510
|
AIB, ABA-inducible BHLH-type transcription factor, ATAIB, ABA-inducible BHLH-type transcription factor, JAM1, JA-associated MYC2-like 1 |
-0.257 |
0.247 |
| 97 |
258245_at |
AT3G29075
|
[glycine-rich protein] |
-0.253 |
-0.029 |
| 98 |
249928_at |
AT5G22250
|
AtCAF1b, CCR4- associated factor 1b, CAF1b, CCR4- associated factor 1b |
-0.251 |
-0.198 |
| 99 |
249886_at |
AT5G22320
|
[Leucine-rich repeat (LRR) family protein] |
-0.233 |
-0.072 |
| 100 |
247954_at |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.229 |
-0.065 |
| 101 |
252199_at |
AT3G50270
|
[HXXXD-type acyl-transferase family protein] |
-0.229 |
-0.025 |
| 102 |
258941_at |
AT3G09940
|
ATMDAR3, ARABIDOPSIS THALIANA MONODEHYDROASCORBATE REDUCTASE 3, MDAR2, MONODEHYDROASCORBATE REDUCTASE 2, MDAR3, MONODEHYDROASCORBATE REDUCTASE 3, MDHAR, monodehydroascorbate reductase |
-0.226 |
0.026 |
| 103 |
255874_at |
AT2G40550
|
ETG1, E2F target gene 1 |
-0.215 |
-0.091 |
| 104 |
251643_at |
AT3G57550
|
AGK2, guanylate kinase, GK-2, GUANYLATE KINAS 2 |
-0.212 |
-0.083 |
| 105 |
248856_at |
AT5G46620
|
unknown |
-0.212 |
0.018 |
| 106 |
248971_at |
AT5G45000
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
-0.210 |
0.096 |
| 107 |
260083_at |
AT1G63220
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-0.208 |
-0.048 |
| 108 |
257923_at |
AT3G23160
|
unknown |
-0.206 |
0.038 |
| 109 |
253162_at |
AT4G35630
|
PSAT1, phosphoserine aminotransferase 1 |
-0.205 |
-0.026 |
| 110 |
250179_at |
AT5G14440
|
[Surfeit locus protein 2 (SURF2)] |
-0.203 |
-0.155 |
| 111 |
252081_at |
AT3G51910
|
AT-HSFA7A, ARABIDOPSIS THALIANA HEAT SHOCK TRANSCRIPTION FACTOR A7A, HSFA7A, heat shock transcription factor A7A |
-0.202 |
-0.078 |
| 112 |
266072_at |
AT2G18700
|
ATTPS11, trehalose phosphatase/synthase 11, ATTPSB, TPS11, trehalose phosphatase/synthase 11, TPS11, TREHALOSE-6-PHOSPHATE SYNTHASE 11 |
-0.201 |
0.058 |
| 113 |
250264_at |
AT5G12890
|
[UDP-Glycosyltransferase superfamily protein] |
-0.194 |
-0.096 |
| 114 |
257321_at |
ATMG01130
|
ORF106F |
-0.193 |
-0.033 |
| 115 |
259749_at |
AT1G71100
|
RSW10, RADIAL SWELLING 10 |
-0.193 |
0.122 |
| 116 |
254432_at |
AT4G20830
|
[FAD-binding Berberine family protein] |
-0.192 |
0.030 |
| 117 |
262383_at |
AT1G72940
|
[Toll-Interleukin-Resistance (TIR) domain-containing protein] |
-0.188 |
-0.005 |
| 118 |
263807_at |
AT2G04400
|
[Aldolase-type TIM barrel family protein] |
-0.184 |
0.046 |
| 119 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.178 |
-0.476 |
| 120 |
258278_at |
AT3G26860
|
[Plant self-incompatibility protein S1 family] |
-0.175 |
0.086 |
| 121 |
265627_at |
AT2G27285
|
[Coiled-coil domain-containing protein 55 (DUF2040)] |
-0.163 |
0.080 |
| 122 |
266802_at |
AT2G22900
|
[Galactosyl transferase GMA12/MNN10 family protein] |
-0.163 |
-0.070 |
| 123 |
260784_at |
AT1G06180
|
ATMYB13, myb domain protein 13, ATMYBLFGN, MYB13, myb domain protein 13 |
-0.162 |
0.077 |
| 124 |
264756_at |
AT1G61370
|
[S-locus lectin protein kinase family protein] |
-0.162 |
0.024 |
| 125 |
262769_at |
AT1G13180
|
ARP3, ACTIN-RELATED PROTEIN 3, ATARP3, ARABIDOPSIS THALIANA ACTIN-RELATED PROTEIN 3, DIS1, DISTORTED TRICHOMES 1 |
-0.162 |
0.121 |