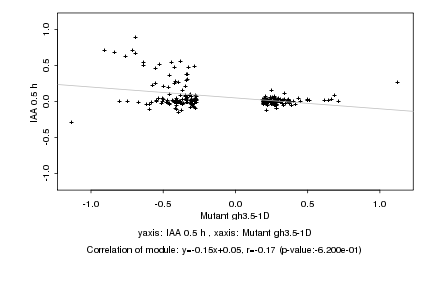
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant gh3.5-1D |
Log2 signal ratio
IAA 0.5 h |
| 1 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
1.121 |
0.274 |
| 2 |
265918_at |
AT2G15090
|
KCS8, 3-ketoacyl-CoA synthase 8 |
0.709 |
0.011 |
| 3 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
0.684 |
0.088 |
| 4 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
0.662 |
0.037 |
| 5 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.640 |
0.024 |
| 6 |
248888_at |
AT5G46240
|
AtKAT1, KAT1, potassium channel in Arabidopsis thaliana 1 |
0.610 |
0.016 |
| 7 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.512 |
0.023 |
| 8 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.498 |
0.036 |
| 9 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.496 |
0.017 |
| 10 |
249115_at |
AT5G43810
|
AGO10, ARGONAUTE 10, PNH, PINHEAD, ZLL, ZWILLE |
0.449 |
0.011 |
| 11 |
261428_at |
AT1G18870
|
ATICS2, ARABIDOPSIS ISOCHORISMATE SYNTHASE 2, ICS2, isochorismate synthase 2 |
0.433 |
0.047 |
| 12 |
264471_at |
AT1G67120
|
AtMDN1, MDN1, MIDASIN 1 |
0.410 |
-0.034 |
| 13 |
265119_at |
AT1G62570
|
FMO GS-OX4, flavin-monooxygenase glucosinolate S-oxygenase 4 |
0.398 |
0.013 |
| 14 |
262464_at |
AT1G50280
|
[Phototropic-responsive NPH3 family protein] |
0.383 |
-0.050 |
| 15 |
254595_at |
AT4G18960
|
AG, AGAMOUS |
0.378 |
0.012 |
| 16 |
256199_at |
AT1G58250
|
HPS4, hypersensitive to Pi starvation 4, SAB, SABRE |
0.374 |
-0.025 |
| 17 |
251432_at |
AT3G59820
|
AtLETM1, LETM1, leucine zipper-EF-hand-containing transmembrane protein 1 |
0.374 |
-0.006 |
| 18 |
265817_at |
AT2G18050
|
HIS1-3, histone H1-3 |
0.371 |
0.009 |
| 19 |
251080_at |
AT5G02010
|
ATROPGEF7, ROPGEF7, ROP (rho of plants) guanine nucleotide exchange factor 7 |
0.368 |
0.013 |
| 20 |
251791_at |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
0.366 |
0.033 |
| 21 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.365 |
-0.012 |
| 22 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.357 |
0.028 |
| 23 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
0.356 |
0.003 |
| 24 |
255836_at |
AT2G33440
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
0.346 |
-0.014 |
| 25 |
248352_at |
AT5G52300
|
LTI65, LOW-TEMPERATURE-INDUCED 65, RD29B, RESPONSIVE TO DESSICATION 29B |
0.334 |
0.114 |
| 26 |
259149_at |
AT3G10340
|
PAL4, phenylalanine ammonia-lyase 4 |
0.333 |
0.029 |
| 27 |
255128_at |
AT4G08310
|
unknown |
0.326 |
-0.042 |
| 28 |
248427_at |
AT5G51750
|
ATSBT1.3, subtilase 1.3, SBT1.3, subtilase 1.3 |
0.326 |
0.028 |
| 29 |
245404_at |
AT4G17610
|
[tRNA/rRNA methyltransferase (SpoU) family protein] |
0.325 |
-0.021 |
| 30 |
261557_at |
AT1G63640
|
[P-loop nucleoside triphosphate hydrolases superfamily protein with CH (Calponin Homology) domain] |
0.322 |
0.022 |
| 31 |
256021_at |
AT1G58270
|
ZW9 |
0.315 |
0.010 |
| 32 |
254506_at |
AT4G20140
|
GSO1, GASSHO1 |
0.309 |
-0.011 |
| 33 |
257516_at |
AT1G69040
|
ACR4, ACT domain repeat 4 |
0.307 |
0.035 |
| 34 |
254073_at |
AT4G25500
|
At-RS40, arginine/serine-rich splicing factor 40, AT-SRP40, ATRSP35, ARABIDOPSIS THALIANA ARGININE/SERINE-RICH SPLICING FACTOR 35, ATRSP40, RS40, arginine/serine-rich splicing factor 40, RSP35, arginine/serine-rich splicing factor 35 |
0.295 |
0.048 |
| 35 |
257024_at |
AT3G19100
|
[Protein kinase superfamily protein] |
0.292 |
0.007 |
| 36 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
0.290 |
0.018 |
| 37 |
264858_at |
AT1G24190
|
ATSIN3, ARABIDOPSIS THALIANA SIN3 HOMOLOG, SIN3, ARABIDOPSIS THALIANA SIN3 HOMOLOG, SNL3, SIN3-like 3 |
0.286 |
0.033 |
| 38 |
254526_at |
AT4G19600
|
CYCT1;4 |
0.283 |
0.017 |
| 39 |
248082_at |
AT5G55400
|
[Actin binding Calponin homology (CH) domain-containing protein] |
0.282 |
-0.035 |
| 40 |
252207_at |
AT3G50370
|
unknown |
0.281 |
-0.006 |
| 41 |
251278_at |
AT3G61780
|
emb1703, embryo defective 1703 |
0.281 |
-0.038 |
| 42 |
262199_at |
AT1G53800
|
unknown |
0.281 |
-0.042 |
| 43 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.280 |
-0.094 |
| 44 |
264987_at |
AT1G27030
|
unknown |
0.279 |
0.012 |
| 45 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.279 |
-0.085 |
| 46 |
264750_at |
AT1G22870
|
[Protein kinase family protein with ARM repeat domain] |
0.275 |
0.010 |
| 47 |
255943_at |
AT1G22370
|
AtUGT85A5, UDP-glucosyl transferase 85A5, UGT85A5, UDP-glucosyl transferase 85A5 |
0.275 |
0.029 |
| 48 |
265807_at |
AT2G17990
|
unknown |
0.272 |
0.014 |
| 49 |
250528_at |
AT5G08600
|
[U3 ribonucleoprotein (Utp) family protein] |
0.271 |
-0.020 |
| 50 |
264338_at |
AT1G70300
|
KUP6, K+ uptake permease 6 |
0.271 |
-0.048 |
| 51 |
264211_at |
AT1G22770
|
FB, GI, GIGANTEA |
0.268 |
-0.044 |
| 52 |
258971_at |
AT3G01990
|
ACR6, ACT domain repeat 6 |
0.268 |
-0.044 |
| 53 |
256394_at |
AT3G06290
|
AtSAC3B, SAC3B, yeast Sac3 homolog B |
0.267 |
0.071 |
| 54 |
255625_at |
AT4G01120
|
ATBZIP54, BASIC REGION/LEUCINE ZIPPER MOTIF 5, GBF2, G-box binding factor 2 |
0.266 |
0.046 |
| 55 |
261421_at |
AT1G18840
|
IQD30, IQ-domain 30 |
0.264 |
0.063 |
| 56 |
245645_at |
AT1G24764
|
ATMAP70-2, microtubule-associated proteins 70-2, MAP70-2, microtubule-associated proteins 70-2 |
0.264 |
0.021 |
| 57 |
264060_at |
AT2G27980
|
[Acyl-CoA N-acyltransferase with RING/FYVE/PHD-type zinc finger domain] |
0.262 |
-0.020 |
| 58 |
246829_at |
AT5G26570
|
ATGWD3, OK1, PWD, PHOSPHOGLUCAN WATER DIKINASE |
0.262 |
-0.011 |
| 59 |
250685_at |
AT5G06670
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.260 |
0.010 |
| 60 |
265312_at |
AT2G20240
|
TRM17, TON1 Recruiting Motif 17 |
0.260 |
-0.015 |
| 61 |
263406_at |
AT2G04160
|
AIR3, AUXIN-INDUCED IN ROOT CULTURES 3 |
0.257 |
0.023 |
| 62 |
261109_at |
AT1G75450
|
ATCKX5, ARABIDOPSIS THALIANA CYTOKININ OXIDASE 5, ATCKX6, CYTOKININ OXIDASE 6, CKX5, cytokinin oxidase 5 |
0.250 |
0.066 |
| 63 |
260731_at |
AT1G17500
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.247 |
0.048 |
| 64 |
247028_at |
AT5G67100
|
ICU2, INCURVATA2 |
0.246 |
-0.011 |
| 65 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
0.244 |
0.014 |
| 66 |
261983_at |
AT1G33770
|
[Protein kinase superfamily protein] |
0.243 |
-0.030 |
| 67 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
0.243 |
0.158 |
| 68 |
253285_at |
AT4G34250
|
KCS16, 3-ketoacyl-CoA synthase 16 |
0.241 |
0.033 |
| 69 |
259945_at |
AT1G71460
|
[Pentatricopeptide repeat (PPR-like) superfamily protein] |
0.240 |
0.009 |
| 70 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.240 |
0.009 |
| 71 |
257085_at |
AT3G20630
|
ATUBP14, PER1, phosphate deficiency root hair defective1, TTN6, TITAN6, UBP14, ubiquitin-specific protease 14 |
0.239 |
0.056 |
| 72 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
0.236 |
0.058 |
| 73 |
246850_at |
AT5G26860
|
LON1, lon protease 1, LON_ARA_ARA |
0.236 |
-0.011 |
| 74 |
253754_at |
AT4G29020
|
[glycine-rich protein] |
0.235 |
0.011 |
| 75 |
264517_at |
AT1G10120
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.233 |
0.020 |
| 76 |
249452_at |
AT5G39500
|
ERMO1, ENDOPLASMIC RETICULUM MORPHOLOGY 1, GNL1, GNOM-like 1 |
0.222 |
-0.013 |
| 77 |
264787_at |
AT2G17840
|
ERD7, EARLY-RESPONSIVE TO DEHYDRATION 7 |
0.222 |
0.020 |
| 78 |
249871_at |
AT5G23110
|
[Zinc finger, C3HC4 type (RING finger) family protein] |
0.221 |
-0.009 |
| 79 |
258994_at |
AT3G08850
|
ATRAPTOR1B, RAPTOR1, RAPTOR1B |
0.220 |
0.010 |
| 80 |
264818_at |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
0.219 |
0.036 |
| 81 |
265896_at |
AT2G25660
|
emb2410, embryo defective 2410 |
0.218 |
-0.049 |
| 82 |
264510_at |
AT1G09530
|
PAP3, PHYTOCHROME-ASSOCIATED PROTEIN 3, PIF3, phytochrome interacting factor 3, POC1, PHOTOCURRENT 1 |
0.218 |
0.023 |
| 83 |
253263_at |
AT4G34000
|
ABF3, abscisic acid responsive elements-binding factor 3, AtABF3, DPBF5, DC3 PROMOTER-BINDING FACTOR 5 |
0.217 |
-0.012 |
| 84 |
260496_at |
AT2G41700
|
ABCA1, ATP-binding cassette A1, AtABCA1, Arabidopsis thaliana ATP-binding cassette A1 |
0.217 |
-0.012 |
| 85 |
267501_at |
AT2G45540
|
[WD-40 repeat family protein / beige-related] |
0.215 |
0.008 |
| 86 |
250971_at |
AT5G02810
|
APRR7, PRR7, pseudo-response regulator 7 |
0.214 |
-0.121 |
| 87 |
261957_at |
AT1G64660
|
ATMGL, methionine gamma-lyase, MGL, methionine gamma-lyase |
0.213 |
-0.040 |
| 88 |
258338_at |
AT3G16150
|
ASPGB1, asparaginase B1 |
0.212 |
0.013 |
| 89 |
264989_at |
AT1G27200
|
unknown |
0.211 |
0.033 |
| 90 |
245202_at |
AT1G67720
|
[Leucine-rich repeat protein kinase family protein] |
0.210 |
-0.002 |
| 91 |
260032_at |
AT1G68750
|
ATPPC4, phosphoenolpyruvate carboxylase 4, PPC4, phosphoenolpyruvate carboxylase 4 |
0.210 |
0.018 |
| 92 |
262800_at |
AT1G20960
|
emb1507, embryo defective 1507 |
0.208 |
-0.009 |
| 93 |
253224_at |
AT4G34860
|
A/N-InvB, alkaline/neutral invertase B |
0.207 |
-0.020 |
| 94 |
262111_at |
AT1G02960
|
unknown |
0.207 |
0.005 |
| 95 |
256659_at |
AT3G12020
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.206 |
0.070 |
| 96 |
261043_at |
AT1G01220
|
AtFKGP, Arabidopsis thaliana L-fucokinase/GDP-L-fucose pyrophosphorylase, FKGP, L-fucokinase/GDP-L-fucose pyrophosphorylase |
0.205 |
-0.013 |
| 97 |
262701_at |
AT1G16480
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.203 |
0.034 |
| 98 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
0.202 |
0.005 |
| 99 |
250520_at |
AT5G08470
|
EMB2817, EMBRYO DEFECTIVE 2817, PEX1, peroxisome 1 |
0.202 |
0.007 |
| 100 |
265395_at |
AT2G20850
|
SRF1, STRUBBELIG-receptor family 1 |
0.202 |
0.016 |
| 101 |
259287_at |
AT3G11490
|
[rac GTPase activating protein] |
0.200 |
0.014 |
| 102 |
247465_at |
AT5G62190
|
PRH75 |
0.200 |
0.017 |
| 103 |
250585_at |
AT5G07620
|
[Protein kinase superfamily protein] |
0.199 |
0.048 |
| 104 |
253603_at |
AT4G30935
|
ATWRKY32, WRKY32, WRKY DNA-binding protein 32 |
0.198 |
0.046 |
| 105 |
250529_at |
AT5G08610
|
PDE340, PIGMENT DEFECTIVE 340 |
0.197 |
-0.002 |
| 106 |
248841_at |
AT5G46740
|
UBP21, ubiquitin-specific protease 21 |
0.197 |
-0.018 |
| 107 |
258901_at |
AT3G05640
|
[Protein phosphatase 2C family protein] |
0.197 |
0.013 |
| 108 |
262784_at |
AT1G10760
|
GWD, GWD1, SEX1, STARCH EXCESS 1, SOP, SOP1 |
0.196 |
-0.009 |
| 109 |
253871_at |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
0.196 |
0.018 |
| 110 |
261909_at |
AT1G80680
|
MOS3, MODIFIER OF SNC1,3, NUP96, PRE, PRECOCIOUS, SAR3, SUPPRESSOR OF AUXIN RESISTANCE 3 |
0.196 |
0.010 |
| 111 |
266380_at |
AT2G14680
|
MEE13, maternal effect embryo arrest 13 |
0.193 |
-0.008 |
| 112 |
254269_at |
AT4G23050
|
[PAS domain-containing protein tyrosine kinase family protein] |
0.193 |
0.007 |
| 113 |
257193_at |
AT3G13160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.192 |
0.011 |
| 114 |
247004_at |
AT5G67570
|
DG1, DELAYED GREENING 1, EMB1408, embryo defective 1408, EMB246, EMBRYO DEFECTIVE 246 |
0.191 |
0.028 |
| 115 |
245210_at |
AT5G12350
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
0.190 |
0.003 |
| 116 |
246232_at |
AT4G36630
|
EMB2754, EMBRYO DEFECTIVE 2754 |
0.190 |
-0.019 |
| 117 |
251975_at |
AT3G53230
|
AtCDC48B, cell division cycle 48B |
0.189 |
-0.015 |
| 118 |
252362_at |
AT3G48500
|
PDE312, PIGMENT DEFECTIVE 312, PTAC10, PLASTID TRANSCRIPTIONALLY ACTIVE 10, TAC10 |
0.188 |
-0.028 |
| 119 |
262066_at |
AT1G79950
|
[RAD3-like DNA-binding helicase protein] |
0.188 |
0.036 |
| 120 |
264389_at |
AT1G11960
|
[ERD (early-responsive to dehydration stress) family protein] |
0.187 |
0.029 |
| 121 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-1.140 |
-0.280 |
| 122 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.913 |
0.716 |
| 123 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.844 |
0.694 |
| 124 |
250764_at |
AT5G05960
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.809 |
0.004 |
| 125 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.766 |
0.638 |
| 126 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
-0.754 |
0.004 |
| 127 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.719 |
0.712 |
| 128 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.697 |
0.894 |
| 129 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.696 |
0.680 |
| 130 |
255127_at |
AT4G08300
|
UMAMIT17, Usually multiple acids move in and out Transporters 17 |
-0.676 |
-0.011 |
| 131 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.641 |
0.506 |
| 132 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.641 |
0.553 |
| 133 |
247800_at |
AT5G58570
|
unknown |
-0.619 |
-0.031 |
| 134 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.600 |
-0.030 |
| 135 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
-0.598 |
-0.102 |
| 136 |
258791_at |
AT3G04720
|
AtPR4, HEL, HEVEIN-LIKE, PR-4, PR4, pathogenesis-related 4 |
-0.582 |
-0.012 |
| 137 |
246200_at |
AT4G37240
|
unknown |
-0.578 |
0.226 |
| 138 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.558 |
0.470 |
| 139 |
247255_at |
AT5G64780
|
[Uncharacterised conserved protein UCP009193] |
-0.558 |
0.254 |
| 140 |
256537_at |
AT1G33340
|
[ENTH/ANTH/VHS superfamily protein] |
-0.552 |
0.008 |
| 141 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-0.551 |
0.023 |
| 142 |
260831_at |
AT1G06830
|
[Glutaredoxin family protein] |
-0.539 |
0.046 |
| 143 |
248801_at |
AT5G47370
|
HAT2 |
-0.529 |
0.520 |
| 144 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-0.516 |
-0.026 |
| 145 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.510 |
0.009 |
| 146 |
250936_at |
AT5G03120
|
unknown |
-0.506 |
0.055 |
| 147 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.502 |
0.031 |
| 148 |
248509_at |
AT5G50335
|
unknown |
-0.501 |
0.215 |
| 149 |
262947_at |
AT1G75750
|
GASA1, GAST1 protein homolog 1 |
-0.479 |
-0.009 |
| 150 |
248327_at |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.475 |
0.023 |
| 151 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.470 |
0.199 |
| 152 |
252501_at |
AT3G46880
|
unknown |
-0.465 |
-0.026 |
| 153 |
249306_at |
AT5G41400
|
[RING/U-box superfamily protein] |
-0.463 |
0.373 |
| 154 |
249383_at |
AT5G39860
|
BHLH136, BASIC HELIX-LOOP-HELIX PROTEIN 136, BNQ1, BANQUO 1, PRE1, PACLOBUTRAZOL RESISTANCE1 |
-0.458 |
0.110 |
| 155 |
250832_at |
AT5G04950
|
ATNAS1, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 1, NAS1, nicotianamine synthase 1 |
-0.457 |
-0.023 |
| 156 |
259632_at |
AT1G56430
|
ATNAS4, ARABIDOPSIS THALIANA NICOTIANAMINE SYNTHASE 4, NAS4, nicotianamine synthase 4 |
-0.457 |
-0.029 |
| 157 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.447 |
0.555 |
| 158 |
265042_at |
AT1G04040
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
-0.439 |
0.017 |
| 159 |
245891_at |
AT5G09220
|
AAP2, amino acid permease 2 |
-0.433 |
0.007 |
| 160 |
253255_at |
AT4G34760
|
SAUR50, SMALL AUXIN UPREGULATED RNA 50 |
-0.428 |
0.259 |
| 161 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.427 |
0.476 |
| 162 |
267385_at |
AT2G44380
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.426 |
-0.013 |
| 163 |
250901_at |
AT5G03530
|
ATRAB, ATRAB ALPHA, ATRAB18B, ATRABC2A, ARABIDOPSIS THALIANA RAB GTPASE HOMOLOG C2A, RABC2A, RAB GTPase homolog C2A |
-0.420 |
0.023 |
| 164 |
261576_at |
AT1G01070
|
UMAMIT28, Usually multiple acids move in and out Transporters 28 |
-0.420 |
-0.097 |
| 165 |
254065_at |
AT4G25420
|
AT2301, ATGA20OX1, ARABIDOPSIS THALIANA GIBBERELLIN 20-OXIDASE 1, GA20OX1, GA5, GA REQUIRING 5 |
-0.415 |
0.291 |
| 166 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
-0.411 |
-0.103 |
| 167 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.411 |
-0.019 |
| 168 |
258349_at |
AT3G17609
|
HYH, HY5-homolog |
-0.409 |
-0.048 |
| 169 |
261700_at |
AT1G32690
|
unknown |
-0.409 |
0.044 |
| 170 |
265722_at |
AT2G40100
|
LHCB4.3, light harvesting complex photosystem II |
-0.408 |
-0.000 |
| 171 |
259661_at |
AT1G55265
|
unknown |
-0.405 |
0.005 |
| 172 |
263133_at |
AT1G78450
|
[SOUL heme-binding family protein] |
-0.405 |
0.016 |
| 173 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
-0.403 |
0.012 |
| 174 |
264071_at |
AT2G27920
|
SCPL51, serine carboxypeptidase-like 51 |
-0.402 |
-0.013 |
| 175 |
245038_at |
AT2G26560
|
PLA IIA, PHOSPHOLIPASE A 2A, PLA2A, phospholipase A 2A, PLAII alpha, PLP2, PATATIN-LIKE PROTEIN 2, PLP2, PHOSPHOLIPASE A 2A |
-0.401 |
-0.144 |
| 176 |
247252_at |
AT5G64770
|
CLEL 9, CLE-like 9, GLV2, GOLVEN 2, RGF9, root meristem growth factor 9 |
-0.400 |
0.277 |
| 177 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
-0.400 |
-0.009 |
| 178 |
249136_at |
AT5G43180
|
unknown |
-0.393 |
0.005 |
| 179 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
-0.388 |
-0.005 |
| 180 |
252220_at |
AT3G49940
|
LBD38, LOB domain-containing protein 38 |
-0.387 |
-0.014 |
| 181 |
254319_at |
AT4G22560
|
unknown |
-0.386 |
0.094 |
| 182 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.385 |
0.567 |
| 183 |
245143_at |
AT2G45450
|
ZPR1, LITTLE ZIPPER 1 |
-0.382 |
0.004 |
| 184 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.382 |
-0.007 |
| 185 |
262902_x_at |
AT1G59930
|
[MADS-box family protein] |
-0.379 |
-0.114 |
| 186 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.371 |
0.164 |
| 187 |
249894_at |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-0.370 |
0.033 |
| 188 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
-0.368 |
-0.031 |
| 189 |
254041_at |
AT4G25830
|
[Uncharacterised protein family (UPF0497)] |
-0.361 |
-0.019 |
| 190 |
252563_at |
AT3G45970
|
ATEXLA1, expansin-like A1, ATEXPL1, ATHEXP BETA 2.1, EXLA1, expansin-like A1, EXPL1, EXPANSIN L1 |
-0.350 |
0.094 |
| 191 |
255511_at |
AT4G02075
|
PIT1, pitchoun 1 |
-0.350 |
0.040 |
| 192 |
264025_at |
AT2G21050
|
LAX2, like AUXIN RESISTANT 2 |
-0.349 |
0.081 |
| 193 |
252965_at |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.347 |
0.215 |
| 194 |
248205_at |
AT5G54300
|
unknown |
-0.344 |
-0.008 |
| 195 |
259272_at |
AT3G01290
|
AtHIR2, HIR2, hypersensitive induced reaction 2 |
-0.342 |
0.055 |
| 196 |
249332_at |
AT5G40980
|
unknown |
-0.342 |
0.027 |
| 197 |
247474_at |
AT5G62280
|
unknown |
-0.340 |
0.306 |
| 198 |
257766_at |
AT3G23030
|
IAA2, indole-3-acetic acid inducible 2 |
-0.340 |
0.384 |
| 199 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
-0.340 |
0.033 |
| 200 |
249167_at |
AT5G42860
|
unknown |
-0.338 |
0.000 |
| 201 |
245362_at |
AT4G17460
|
HAT1, JAB, JAIBA |
-0.337 |
0.313 |
| 202 |
255129_at |
AT4G08290
|
UMAMIT20, Usually multiple acids move in and out Transporters 20 |
-0.334 |
-0.009 |
| 203 |
263931_at |
AT2G36220
|
unknown |
-0.332 |
0.387 |
| 204 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.332 |
0.056 |
| 205 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.325 |
0.478 |
| 206 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-0.321 |
0.097 |
| 207 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
-0.315 |
-0.076 |
| 208 |
264179_at |
AT1G02180
|
[ferredoxin-related] |
-0.315 |
-0.016 |
| 209 |
262263_at |
AT1G70940
|
ATPIN3, ARABIDOPSIS PIN-FORMED 3, PIN3, PIN-FORMED 3 |
-0.314 |
0.100 |
| 210 |
262290_at |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-0.312 |
0.025 |
| 211 |
263840_at |
AT2G36885
|
unknown |
-0.311 |
-0.032 |
| 212 |
257242_at |
AT3G24220
|
ATNCED6, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 6, NCED6, nine-cis-epoxycarotenoid dioxygenase 6 |
-0.310 |
0.035 |
| 213 |
245574_at |
AT4G14750
|
IQD19, IQ-domain 19 |
-0.309 |
0.013 |
| 214 |
267590_at |
AT2G39700
|
ATEXP4, ATEXPA4, expansin A4, ATHEXP ALPHA 1.6, EXPA4, expansin A4 |
-0.309 |
0.023 |
| 215 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.309 |
0.074 |
| 216 |
249867_at |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
-0.303 |
-0.014 |
| 217 |
265984_at |
AT2G24210
|
TPS10, terpene synthase 10 |
-0.300 |
-0.030 |
| 218 |
248686_at |
AT5G48540
|
[receptor-like protein kinase-related family protein] |
-0.300 |
0.018 |
| 219 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
-0.292 |
-0.058 |
| 220 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.289 |
0.500 |
| 221 |
253401_at |
AT4G32870
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.289 |
-0.014 |
| 222 |
257793_at |
AT3G26960
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.288 |
0.040 |
| 223 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.287 |
0.012 |
| 224 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
-0.286 |
0.025 |
| 225 |
254387_at |
AT4G21850
|
ATMSRB9, methionine sulfoxide reductase B9, MSRB9, methionine sulfoxide reductase B9 |
-0.286 |
0.018 |
| 226 |
262698_at |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
-0.285 |
-0.082 |
| 227 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.283 |
0.042 |
| 228 |
261881_at |
AT1G80760
|
NIP6, NIP6;1, NOD26-like intrinsic protein 6;1, NLM7 |
-0.283 |
0.035 |
| 229 |
255417_at |
AT4G03190
|
AFB1, AUXIN SIGNALING F BOX PROTEIN 1, ATGRH1, GRH1, GRR1-like protein 1 |
-0.283 |
0.024 |
| 230 |
263318_at |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.282 |
0.023 |
| 231 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.281 |
0.045 |
| 232 |
258385_at |
AT3G15510
|
ANAC056, Arabidopsis NAC domain containing protein 56, ATNAC2, NAC domain containing protein 2, NAC2, NAC domain containing protein 2, NARS1, NAC-REGULATED SEED MORPHOLOGY 1 |
-0.280 |
-0.093 |
| 233 |
251640_at |
AT3G57450
|
unknown |
-0.278 |
0.085 |
| 234 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.277 |
0.035 |
| 235 |
252539_at |
AT3G45730
|
unknown |
-0.277 |
0.051 |
| 236 |
263498_at |
AT2G42610
|
LSH10, LIGHT SENSITIVE HYPOCOTYLS 10 |
-0.276 |
0.018 |
| 237 |
254573_at |
AT4G19420
|
[Pectinacetylesterase family protein] |
-0.276 |
-0.001 |
| 238 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.275 |
0.038 |
| 239 |
248246_at |
AT5G53200
|
TRY, TRIPTYCHON |
-0.274 |
0.079 |