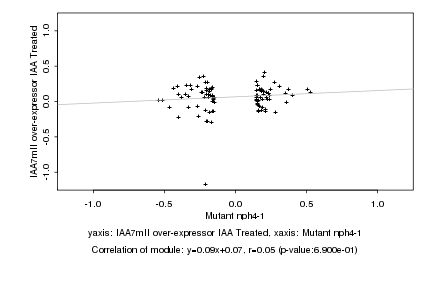
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant nph4-1 |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.524 |
0.139 |
| 2 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.503 |
0.180 |
| 3 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.396 |
0.092 |
| 4 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.372 |
0.184 |
| 5 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.356 |
-0.012 |
| 6 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.351 |
0.127 |
| 7 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.306 |
0.216 |
| 8 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.276 |
-0.146 |
| 9 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.269 |
0.271 |
| 10 |
245550_at |
AT4G15330
|
CYP705A1, cytochrome P450, family 705, subfamily A, polypeptide 1 |
0.240 |
0.178 |
| 11 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
0.239 |
0.097 |
| 12 |
257089_at |
AT3G20520
|
GDPDL5, Glycerophosphodiester phosphodiesterase (GDPD) like 5, SVL3, SHV3-like 3 |
0.237 |
0.032 |
| 13 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.230 |
0.117 |
| 14 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.226 |
0.123 |
| 15 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.221 |
0.031 |
| 16 |
260416_at |
AT1G69670
|
ATCUL3B, ARABIDOPSIS THALIANA CULLIN 3B, CUL3B, cullin 3B |
0.217 |
0.064 |
| 17 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.214 |
0.140 |
| 18 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.208 |
-0.130 |
| 19 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.205 |
-0.111 |
| 20 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.198 |
0.424 |
| 21 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.195 |
0.104 |
| 22 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.195 |
0.149 |
| 23 |
259282_at |
AT3G11430
|
ATGPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5, GPAT5, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 5 |
0.192 |
0.362 |
| 24 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.189 |
0.159 |
| 25 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
0.188 |
0.034 |
| 26 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.187 |
-0.077 |
| 27 |
250823_at |
AT5G05180
|
unknown |
0.178 |
-0.120 |
| 28 |
262811_at |
AT1G11700
|
unknown |
0.178 |
0.176 |
| 29 |
262324_at |
AT1G64170
|
ATCHX16, cation/H+ exchanger 16, CHX16, cation/H+ exchanger 16 |
0.177 |
0.151 |
| 30 |
263825_at |
AT2G40370
|
LAC5, laccase 5 |
0.176 |
0.062 |
| 31 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.175 |
0.167 |
| 32 |
260234_at |
AT1G74460
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.172 |
0.167 |
| 33 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.169 |
0.175 |
| 34 |
267036_at |
AT2G38465
|
unknown |
0.166 |
-0.066 |
| 35 |
246331_at |
AT3G44820
|
[Phototropic-responsive NPH3 family protein] |
0.163 |
0.173 |
| 36 |
265083_at |
AT1G03820
|
unknown |
0.161 |
-0.052 |
| 37 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.159 |
-0.134 |
| 38 |
258254_at |
AT3G26782
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.159 |
0.016 |
| 39 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
0.156 |
-0.116 |
| 40 |
249320_at |
AT5G40910
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.155 |
-0.017 |
| 41 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.155 |
0.234 |
| 42 |
249881_at |
AT5G23190
|
CYP86B1, cytochrome P450, family 86, subfamily B, polypeptide 1 |
0.155 |
-0.036 |
| 43 |
253373_at |
AT4G33150
|
LKR, LKR/SDH, LYSINE-KETOGLUTARATE REDUCTASE/SACCHAROPINE DEHYDROGENASE, SDH, SACCHAROPINE DEHYDROGENASE |
0.155 |
0.090 |
| 44 |
249953_at |
AT5G18960
|
FRS12, FAR1-related sequence 12 |
0.151 |
0.011 |
| 45 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.150 |
0.230 |
| 46 |
250239_at |
AT5G13580
|
ABCG6, ATP-binding cassette G6 |
0.148 |
0.098 |
| 47 |
249576_at |
AT5G37690
|
[SGNH hydrolase-type esterase superfamily protein] |
0.147 |
0.289 |
| 48 |
259992_at |
AT1G67970
|
AT-HSFA8, HSFA8, heat shock transcription factor A8 |
0.147 |
0.062 |
| 49 |
260611_at |
AT2G43670
|
[Carbohydrate-binding X8 domain superfamily protein] |
0.145 |
0.041 |
| 50 |
261402_at |
AT1G79670
|
RFO1, RESISTANCE TO FUSARIUM OXYSPORUM 1, WAKL22 |
0.145 |
0.018 |
| 51 |
254777_at |
AT4G12960
|
AtGILT, GILT, gamma-interferon-responsive lysosomal thiol reductase |
0.145 |
0.156 |
| 52 |
245971_at |
AT5G20730
|
ARF7, AUXIN RESPONSE FACTOR 7, BIP, BIPOSTO, IAA21, indole-3-acetic acid inducible 21, IAA23, indole-3-acetic acid inducible 23, IAA25, indole-3-acetic acid inducible 25, MSG1, MASSUGU 1, NPH4, NON-PHOTOTROPHIC HYPOCOTYL, TIR5, TRANSPORT INHIBITOR RESPONSE 5 |
-0.548 |
0.025 |
| 53 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.514 |
0.018 |
| 54 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.471 |
-0.072 |
| 55 |
247878_at |
AT5G57760
|
unknown |
-0.439 |
0.185 |
| 56 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.409 |
0.214 |
| 57 |
248509_at |
AT5G50335
|
unknown |
-0.408 |
0.110 |
| 58 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.407 |
-0.220 |
| 59 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
-0.387 |
0.063 |
| 60 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.353 |
0.113 |
| 61 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.347 |
0.230 |
| 62 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.336 |
-0.072 |
| 63 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.335 |
0.077 |
| 64 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.317 |
0.228 |
| 65 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.316 |
0.182 |
| 66 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.274 |
-0.057 |
| 67 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.268 |
0.219 |
| 68 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.262 |
-0.203 |
| 69 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.255 |
0.340 |
| 70 |
254609_at |
AT4G18970
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.242 |
0.135 |
| 71 |
259015_at |
AT3G07350
|
unknown |
-0.234 |
0.131 |
| 72 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.229 |
0.365 |
| 73 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.221 |
0.070 |
| 74 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.217 |
-0.113 |
| 75 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.216 |
0.270 |
| 76 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.214 |
-1.160 |
| 77 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.210 |
0.191 |
| 78 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.209 |
0.161 |
| 79 |
267230_at |
AT2G44080
|
ARL, ARGOS-like |
-0.207 |
0.108 |
| 80 |
259927_at |
AT1G75100
|
JAC1, J-domain protein required for chloroplast accumulation response 1 |
-0.207 |
-0.273 |
| 81 |
260856_at |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-0.202 |
0.274 |
| 82 |
252539_at |
AT3G45730
|
unknown |
-0.198 |
-0.282 |
| 83 |
247880_at |
AT5G57780
|
P1R1, P1R1 |
-0.196 |
0.060 |
| 84 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.196 |
0.102 |
| 85 |
259163_at |
AT3G01490
|
[Protein kinase superfamily protein] |
-0.188 |
-0.147 |
| 86 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.188 |
0.184 |
| 87 |
251491_at |
AT3G59480
|
[pfkB-like carbohydrate kinase family protein] |
-0.185 |
0.146 |
| 88 |
256266_at |
AT3G12320
|
LNK3, night light-inducible and clock-regulated 3 |
-0.182 |
0.099 |
| 89 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.175 |
-0.291 |
| 90 |
256285_at |
AT3G12510
|
[MADS-box family protein] |
-0.171 |
0.188 |
| 91 |
250049_at |
AT5G17780
|
[alpha/beta-Hydrolases superfamily protein] |
-0.169 |
0.179 |
| 92 |
250472_at |
AT5G10210
|
unknown |
-0.168 |
0.087 |
| 93 |
259275_at |
AT3G01060
|
unknown |
-0.166 |
0.084 |
| 94 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.165 |
0.009 |
| 95 |
252167_at |
AT3G50560
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.163 |
0.208 |
| 96 |
246919_at |
AT5G25460
|
DGR2, DUF642 L-GalL responsive gene 2 |
-0.163 |
-0.135 |
| 97 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.160 |
0.055 |
| 98 |
264395_at |
AT1G12070
|
[Immunoglobulin E-set superfamily protein] |
-0.160 |
0.029 |
| 99 |
249035_at |
AT5G44190
|
ATGLK2, GLK2, GOLDEN2-like 2, GPRI2, GBF'S PRO-RICH REGION-INTERACTING FACTOR 2 |
-0.159 |
-0.132 |
| 100 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.154 |
0.068 |
| 101 |
247585_at |
AT5G60680
|
unknown |
-0.150 |
-0.013 |