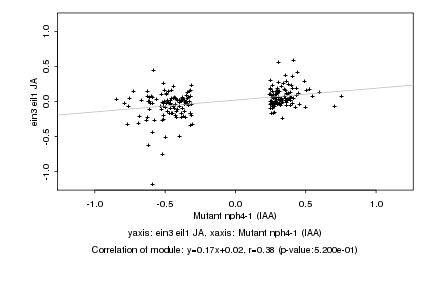
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant nph4-1 (IAA) |
Log2 signal ratio
ein3 eil1 JA |
| 1 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.750 |
0.085 |
| 2 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.703 |
-0.065 |
| 3 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.596 |
0.139 |
| 4 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.547 |
0.073 |
| 5 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.526 |
0.179 |
| 6 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.505 |
0.162 |
| 7 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.497 |
-0.085 |
| 8 |
261221_at |
AT1G19960
|
unknown |
0.489 |
0.290 |
| 9 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.450 |
-0.033 |
| 10 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.442 |
0.119 |
| 11 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.441 |
0.415 |
| 12 |
262811_at |
AT1G11700
|
unknown |
0.428 |
0.098 |
| 13 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.428 |
0.190 |
| 14 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.425 |
-0.076 |
| 15 |
266222_at |
AT2G28780
|
unknown |
0.412 |
0.057 |
| 16 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.406 |
0.596 |
| 17 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.404 |
0.361 |
| 18 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.399 |
0.186 |
| 19 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
0.399 |
-0.014 |
| 20 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.397 |
0.232 |
| 21 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.394 |
0.059 |
| 22 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.387 |
0.016 |
| 23 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.386 |
0.027 |
| 24 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.385 |
0.069 |
| 25 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.385 |
0.141 |
| 26 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
0.385 |
-0.050 |
| 27 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.382 |
0.051 |
| 28 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
0.375 |
0.119 |
| 29 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.372 |
0.256 |
| 30 |
263545_at |
AT2G21560
|
unknown |
0.369 |
0.025 |
| 31 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.368 |
0.034 |
| 32 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
0.367 |
0.101 |
| 33 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.361 |
-0.002 |
| 34 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.359 |
-0.056 |
| 35 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.358 |
0.117 |
| 36 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.357 |
0.300 |
| 37 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.354 |
0.375 |
| 38 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.354 |
0.164 |
| 39 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.352 |
0.019 |
| 40 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.351 |
0.047 |
| 41 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.347 |
0.179 |
| 42 |
259576_at |
AT1G35330
|
[RING/U-box superfamily protein] |
0.342 |
0.085 |
| 43 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.335 |
0.269 |
| 44 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.330 |
-0.011 |
| 45 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.328 |
-0.235 |
| 46 |
246844_at |
AT5G26660
|
ATMYB86, myb domain protein 86, MYB86, myb domain protein 86 |
0.326 |
0.032 |
| 47 |
257074_at |
AT3G19660
|
unknown |
0.326 |
0.002 |
| 48 |
250937_at |
AT5G03230
|
unknown |
0.325 |
0.027 |
| 49 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.324 |
-0.014 |
| 50 |
258252_at |
AT3G15720
|
[Pectin lyase-like superfamily protein] |
0.322 |
0.039 |
| 51 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
0.322 |
0.051 |
| 52 |
257650_at |
AT3G16800
|
[Protein phosphatase 2C family protein] |
0.321 |
0.215 |
| 53 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
0.320 |
0.029 |
| 54 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.317 |
0.022 |
| 55 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.314 |
-0.053 |
| 56 |
248790_at |
AT5G47450
|
ATTIP2;3, ARABIDOPSIS THALIANA TONOPLAST INTRINSIC PROTEIN 2;3, DELTA-TIP3, DELTA-TONOPLAST INTRINSIC PROTEIN 3, TIP2;3, tonoplast intrinsic protein 2;3 |
0.310 |
0.156 |
| 57 |
253515_at |
AT4G31320
|
SAUR37, SMALL AUXIN UPREGULATED RNA 37 |
0.308 |
-0.020 |
| 58 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.308 |
-0.020 |
| 59 |
266596_at |
AT2G46150
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.305 |
0.145 |
| 60 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.304 |
0.042 |
| 61 |
250741_at |
AT5G05790
|
[Duplicated homeodomain-like superfamily protein] |
0.304 |
0.061 |
| 62 |
259388_at |
AT1G13420
|
ATST4B, ARABIDOPSIS THALIANA SULFOTRANSFERASE 4B, ST4B, sulfotransferase 4B |
0.301 |
0.564 |
| 63 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
0.301 |
0.059 |
| 64 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.301 |
0.031 |
| 65 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
0.300 |
0.277 |
| 66 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.298 |
-0.011 |
| 67 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.298 |
0.190 |
| 68 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.297 |
0.161 |
| 69 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
0.296 |
0.124 |
| 70 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.296 |
-0.037 |
| 71 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.296 |
0.142 |
| 72 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.291 |
0.156 |
| 73 |
247582_at |
AT5G60760
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.291 |
0.043 |
| 74 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.290 |
0.111 |
| 75 |
246142_at |
AT5G19970
|
unknown |
0.290 |
0.067 |
| 76 |
262598_at |
AT1G15260
|
unknown |
0.286 |
0.145 |
| 77 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.285 |
-0.026 |
| 78 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
0.284 |
0.004 |
| 79 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.284 |
0.069 |
| 80 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.280 |
0.111 |
| 81 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.279 |
-0.002 |
| 82 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
0.275 |
-0.005 |
| 83 |
245258_at |
AT4G15340
|
04C11, ATPEN1, pentacyclic triterpene synthase 1, PEN1, pentacyclic triterpene synthase 1 |
0.275 |
-0.067 |
| 84 |
266156_at |
AT2G28110
|
FRA8, FRAGILE FIBER 8, IRX7, IRREGULAR XYLEM 7 |
0.274 |
-0.145 |
| 85 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
0.272 |
-0.032 |
| 86 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.272 |
0.070 |
| 87 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
0.270 |
0.102 |
| 88 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
0.269 |
-0.066 |
| 89 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.266 |
0.044 |
| 90 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.266 |
0.108 |
| 91 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.266 |
0.064 |
| 92 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.266 |
-0.164 |
| 93 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
0.265 |
-0.047 |
| 94 |
254693_at |
AT4G17880
|
MYC4 |
0.263 |
0.065 |
| 95 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.262 |
-0.095 |
| 96 |
246911_at |
AT5G25810
|
tny, TINY |
0.262 |
-0.013 |
| 97 |
258078_at |
AT3G25870
|
unknown |
0.260 |
-0.005 |
| 98 |
264052_at |
AT2G22330
|
CYP79B3, cytochrome P450, family 79, subfamily B, polypeptide 3 |
0.259 |
0.153 |
| 99 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.258 |
-0.021 |
| 100 |
251298_at |
AT3G62040
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.258 |
0.234 |
| 101 |
259832_at |
AT1G69580
|
[Homeodomain-like superfamily protein] |
0.257 |
-0.064 |
| 102 |
257900_at |
AT3G28420
|
[Putative membrane lipoprotein] |
0.256 |
0.010 |
| 103 |
261453_at |
AT1G21130
|
IGMT4, indole glucosinolate O-methyltransferase 4 |
0.255 |
-0.164 |
| 104 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.253 |
-0.028 |
| 105 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.252 |
0.013 |
| 106 |
262533_at |
AT1G17090
|
unknown |
0.251 |
0.092 |
| 107 |
252180_at |
AT3G50630
|
ICK2, KRP2, KIP-related protein 2 |
0.251 |
-0.026 |
| 108 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.249 |
0.307 |
| 109 |
250438_at |
AT5G10580
|
unknown |
0.247 |
-0.040 |
| 110 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.246 |
-0.088 |
| 111 |
261921_at |
AT1G65900
|
unknown |
0.245 |
0.105 |
| 112 |
253172_at |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
0.244 |
0.192 |
| 113 |
255448_at |
AT4G02810
|
FAF1, FANTASTIC FOUR 1 |
0.244 |
0.103 |
| 114 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.243 |
0.014 |
| 115 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.243 |
0.183 |
| 116 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.242 |
0.109 |
| 117 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.851 |
0.038 |
| 118 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.795 |
-0.024 |
| 119 |
247878_at |
AT5G57760
|
unknown |
-0.772 |
-0.318 |
| 120 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
-0.764 |
-0.060 |
| 121 |
250709_at |
AT5G06080
|
LBD33, LOB domain-containing protein 33 |
-0.756 |
0.045 |
| 122 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
-0.732 |
0.153 |
| 123 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.696 |
-0.313 |
| 124 |
253155_at |
AT4G35720
|
unknown |
-0.684 |
-0.206 |
| 125 |
267250_at |
AT2G23060
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.670 |
0.016 |
| 126 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.638 |
-0.269 |
| 127 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.632 |
0.145 |
| 128 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.631 |
-0.215 |
| 129 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.629 |
0.085 |
| 130 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.624 |
0.006 |
| 131 |
248509_at |
AT5G50335
|
unknown |
-0.623 |
-0.621 |
| 132 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.612 |
0.059 |
| 133 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.612 |
-0.103 |
| 134 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.612 |
0.014 |
| 135 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.608 |
-0.026 |
| 136 |
247179_at |
AT5G65320
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.604 |
0.081 |
| 137 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.594 |
0.060 |
| 138 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.593 |
-0.438 |
| 139 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.593 |
-1.171 |
| 140 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.591 |
-0.020 |
| 141 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.584 |
0.456 |
| 142 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.578 |
-0.270 |
| 143 |
251377_at |
AT3G60650
|
unknown |
-0.565 |
0.036 |
| 144 |
245971_at |
AT5G20730
|
ARF7, AUXIN RESPONSE FACTOR 7, BIP, BIPOSTO, IAA21, indole-3-acetic acid inducible 21, IAA23, indole-3-acetic acid inducible 23, IAA25, indole-3-acetic acid inducible 25, MSG1, MASSUGU 1, NPH4, NON-PHOTOTROPHIC HYPOCOTYL, TIR5, TRANSPORT INHIBITOR RESPONSE 5 |
-0.540 |
-0.060 |
| 145 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.532 |
-0.103 |
| 146 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.531 |
0.111 |
| 147 |
251293_at |
AT3G61930
|
unknown |
-0.525 |
-0.001 |
| 148 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.524 |
-0.749 |
| 149 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.523 |
-0.262 |
| 150 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.518 |
-0.059 |
| 151 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.518 |
-0.244 |
| 152 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.514 |
-0.186 |
| 153 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-0.513 |
-0.015 |
| 154 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.512 |
0.263 |
| 155 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.509 |
0.001 |
| 156 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.508 |
0.166 |
| 157 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.504 |
-0.501 |
| 158 |
252204_at |
AT3G50340
|
unknown |
-0.501 |
-0.076 |
| 159 |
251612_at |
AT3G57950
|
unknown |
-0.494 |
0.110 |
| 160 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.494 |
-0.017 |
| 161 |
253047_at |
AT4G37295
|
unknown |
-0.493 |
0.127 |
| 162 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.489 |
0.021 |
| 163 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.486 |
-0.006 |
| 164 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.486 |
-0.132 |
| 165 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.482 |
0.145 |
| 166 |
261651_at |
AT1G27760
|
ATSAT32, SALT-TOLERANCE 32, SAT32, SALT-TOLERANCE 32 |
-0.477 |
-0.001 |
| 167 |
248717_at |
AT5G48175
|
unknown |
-0.473 |
-0.171 |
| 168 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
-0.470 |
-0.023 |
| 169 |
248456_at |
AT5G51380
|
[RNI-like superfamily protein] |
-0.465 |
-0.020 |
| 170 |
266941_at |
AT2G18980
|
[Peroxidase superfamily protein] |
-0.464 |
-0.019 |
| 171 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.463 |
-0.039 |
| 172 |
258878_at |
AT3G03170
|
unknown |
-0.463 |
0.015 |
| 173 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.462 |
-0.095 |
| 174 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.460 |
0.051 |
| 175 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.458 |
0.161 |
| 176 |
263436_at |
AT2G28690
|
unknown |
-0.455 |
-0.169 |
| 177 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.454 |
-0.166 |
| 178 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
-0.442 |
0.217 |
| 179 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.441 |
0.054 |
| 180 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
-0.441 |
-0.069 |
| 181 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
-0.435 |
0.064 |
| 182 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.434 |
-0.090 |
| 183 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.428 |
-0.041 |
| 184 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.428 |
0.049 |
| 185 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.427 |
0.052 |
| 186 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.427 |
-0.186 |
| 187 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
-0.423 |
-0.226 |
| 188 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.423 |
0.040 |
| 189 |
245416_at |
AT4G17350
|
unknown |
-0.422 |
-0.134 |
| 190 |
248623_at |
AT5G49170
|
unknown |
-0.417 |
-0.104 |
| 191 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.415 |
0.023 |
| 192 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.403 |
-0.490 |
| 193 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.403 |
-0.010 |
| 194 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.401 |
0.022 |
| 195 |
247003_at |
AT5G67550
|
unknown |
-0.401 |
-0.062 |
| 196 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-0.398 |
0.024 |
| 197 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.390 |
0.008 |
| 198 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
-0.386 |
-0.162 |
| 199 |
247474_at |
AT5G62280
|
unknown |
-0.385 |
-0.217 |
| 200 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
-0.383 |
0.066 |
| 201 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.382 |
0.028 |
| 202 |
256030_at |
AT1G34110
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.382 |
0.037 |
| 203 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.381 |
0.052 |
| 204 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.378 |
-0.090 |
| 205 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.374 |
-0.203 |
| 206 |
253662_at |
AT4G30080
|
ARF16, auxin response factor 16 |
-0.369 |
0.021 |
| 207 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.367 |
-0.130 |
| 208 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
-0.366 |
-0.040 |
| 209 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.365 |
0.028 |
| 210 |
248548_at |
AT5G50300
|
ATAZG2, ARABIDOPSIS THALIANA AZA-GUANINE RESISTANT2, AZG2, AZA-GUANINE RESISTANT2 |
-0.358 |
-0.009 |
| 211 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.356 |
-0.228 |
| 212 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.354 |
0.095 |
| 213 |
256799_at |
AT3G18560
|
unknown |
-0.352 |
-0.028 |
| 214 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-0.350 |
0.006 |
| 215 |
250145_at |
AT5G14690
|
unknown |
-0.347 |
0.135 |
| 216 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.344 |
0.049 |
| 217 |
257049_at |
AT3G15250
|
unknown |
-0.342 |
-0.025 |
| 218 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
-0.335 |
0.067 |
| 219 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.334 |
-0.047 |
| 220 |
253179_at |
AT4G35200
|
unknown |
-0.333 |
0.008 |
| 221 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
-0.327 |
0.149 |
| 222 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.326 |
0.083 |
| 223 |
249887_at |
AT5G22310
|
unknown |
-0.326 |
-0.171 |
| 224 |
253829_at |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-0.324 |
-0.341 |
| 225 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.323 |
0.073 |
| 226 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.321 |
-0.171 |
| 227 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.321 |
0.164 |
| 228 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.319 |
-0.192 |
| 229 |
255694_at |
AT4G00050
|
UNE10, unfertilized embryo sac 10 |
-0.316 |
-0.036 |
| 230 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.315 |
0.243 |
| 231 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
-0.312 |
-0.323 |