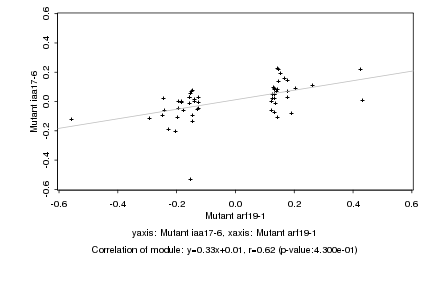
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant arf19-1 |
Log2 signal ratio
Mutant iaa17-6 |
| 1 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.429 |
0.011 |
| 2 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.423 |
0.223 |
| 3 |
260092_at |
AT1G73280
|
scpl3, serine carboxypeptidase-like 3 |
0.259 |
0.112 |
| 4 |
267364_at |
AT2G40080
|
ELF4, EARLY FLOWERING 4 |
0.204 |
0.095 |
| 5 |
256027_at |
AT1G34160
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.189 |
-0.077 |
| 6 |
256563_at |
AT3G29780
|
RALFL27, ralf-like 27 |
0.176 |
0.033 |
| 7 |
266066_at |
AT2G18800
|
ATXTH21, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 21, XTH21, xyloglucan endotransglucosylase/hydrolase 21 |
0.176 |
0.072 |
| 8 |
250500_at |
AT5G09530
|
PELPK1, Pro-Glu-Leu|Ile|Val-Pro-Lys 1, PRP10, proline-rich protein 10 |
0.176 |
0.146 |
| 9 |
265355_at |
AT2G16760
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.166 |
0.159 |
| 10 |
251517_at |
AT3G59370
|
[Vacuolar calcium-binding protein-related] |
0.151 |
0.193 |
| 11 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.146 |
0.219 |
| 12 |
267121_at |
AT2G23540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.145 |
0.141 |
| 13 |
259735_at |
AT1G64405
|
unknown |
0.142 |
-0.109 |
| 14 |
245181_at |
AT5G12420
|
[O-acyltransferase (WSD1-like) family protein] |
0.140 |
0.084 |
| 15 |
249527_at |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
0.140 |
0.227 |
| 16 |
260296_at |
AT1G63750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.139 |
0.075 |
| 17 |
246476_at |
AT5G16730
|
[LOCATED IN: chloroplast] |
0.136 |
-0.007 |
| 18 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
0.132 |
0.054 |
| 19 |
263283_at |
AT2G36090
|
[F-box family protein] |
0.132 |
0.050 |
| 20 |
245630_at |
AT1G25360
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
0.132 |
0.022 |
| 21 |
258178_at |
AT3G21680
|
unknown |
0.131 |
0.088 |
| 22 |
249470_at |
AT5G39350
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.131 |
-0.070 |
| 23 |
264937_at |
AT1G61130
|
SCPL32, serine carboxypeptidase-like 32 |
0.130 |
0.095 |
| 24 |
258013_at |
AT3G19320
|
[Leucine-rich repeat (LRR) family protein] |
0.128 |
0.099 |
| 25 |
266231_at |
AT2G02220
|
ATPSKR1, PHYTOSULFOKIN RECEPTOR 1, PSKR1, phytosulfokin receptor 1 |
0.124 |
0.027 |
| 26 |
266913_at |
AT2G45890
|
ATROPGEF4, RHO GUANYL-NUCLEOTIDE EXCHANGE FACTOR 4, GEF4, guanine exchange factor 4, RHS11, ROOT HAIR SPECIFIC 11, ROPGEF4, ROP (rho of plants) guanine nucleotide exchange factor 4 |
0.123 |
0.052 |
| 27 |
258342_at |
AT3G22800
|
[Leucine-rich repeat (LRR) family protein] |
0.122 |
0.002 |
| 28 |
259562_at |
AT1G21200
|
[sequence-specific DNA binding transcription factors] |
0.122 |
-0.055 |
| 29 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
-0.560 |
-0.119 |
| 30 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-0.295 |
-0.114 |
| 31 |
248311_at |
AT5G52570
|
B2, BCH2, BETA CAROTENOID HYDROXYLASE 2, BETA-OHASE 2, beta-carotene hydroxylase 2, CHY2 |
-0.249 |
-0.089 |
| 32 |
258497_at |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.248 |
0.025 |
| 33 |
258239_at |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
-0.244 |
-0.055 |
| 34 |
261292_at |
AT1G36940
|
unknown |
-0.231 |
-0.188 |
| 35 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.204 |
-0.202 |
| 36 |
259751_at |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-0.199 |
-0.107 |
| 37 |
252412_at |
AT3G47295
|
unknown |
-0.196 |
0.005 |
| 38 |
250561_at |
AT5G08030
|
AtGDPD6, GDPD6, glycerophosphodiester phosphodiesterase 6 |
-0.194 |
-0.041 |
| 39 |
260614_at |
AT1G53390
|
ABCG24, ATP-binding cassette G24 |
-0.186 |
-0.005 |
| 40 |
265111_at |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.185 |
0.003 |
| 41 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.178 |
-0.056 |
| 42 |
250665_at |
AT5G06980
|
LNK4, night light-inducible and clock-regulated 4 |
-0.159 |
0.028 |
| 43 |
248319_at |
AT5G52710
|
[Copper transport protein family] |
-0.158 |
-0.007 |
| 44 |
258809_at |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-0.157 |
0.030 |
| 45 |
267063_at |
AT2G41120
|
unknown |
-0.155 |
0.060 |
| 46 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.153 |
-0.531 |
| 47 |
250472_at |
AT5G10210
|
unknown |
-0.152 |
0.075 |
| 48 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.149 |
-0.135 |
| 49 |
251072_at |
AT5G01740
|
[Nuclear transport factor 2 (NTF2) family protein] |
-0.148 |
-0.091 |
| 50 |
247214_at |
AT5G64850
|
unknown |
-0.147 |
0.082 |
| 51 |
248207_at |
AT5G53970
|
TAT7, tyrosine aminotransferase 7 |
-0.141 |
0.001 |
| 52 |
264195_at |
AT1G22690
|
[Gibberellin-regulated family protein] |
-0.141 |
0.018 |
| 53 |
267529_at |
AT2G45490
|
AtAUR3, ataurora3, AUR3, ataurora3 |
-0.131 |
-0.049 |
| 54 |
266481_at |
AT2G31070
|
TCP10, TCP domain protein 10 |
-0.128 |
-0.003 |
| 55 |
256816_at |
AT3G21400
|
unknown |
-0.127 |
0.030 |
| 56 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.127 |
-0.042 |