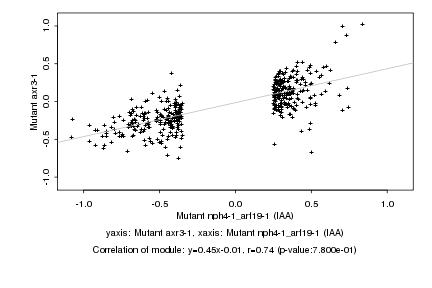
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant nph4-1_arf19-1 (IAA) |
Log2 signal ratio
Mutant axr3-1 |
| 1 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.834 |
1.020 |
| 2 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.743 |
-0.071 |
| 3 |
248978_at |
AT5G45070
|
AtPP2-A8, phloem protein 2-A8, PP2-A8, phloem protein 2-A8 |
0.736 |
0.179 |
| 4 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.727 |
0.874 |
| 5 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
0.702 |
1.000 |
| 6 |
249599_at |
AT5G37990
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.700 |
-0.109 |
| 7 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.681 |
0.083 |
| 8 |
263829_at |
AT2G40435
|
unknown |
0.655 |
0.786 |
| 9 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.622 |
0.413 |
| 10 |
247478_at |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.615 |
0.243 |
| 11 |
261221_at |
AT1G19960
|
unknown |
0.597 |
0.476 |
| 12 |
249174_at |
AT5G42900
|
COR27, cold regulated gene 27 |
0.593 |
0.135 |
| 13 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.578 |
0.456 |
| 14 |
249202_at |
AT5G42580
|
CYP705A12, cytochrome P450, family 705, subfamily A, polypeptide 12 |
0.564 |
0.101 |
| 15 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.564 |
0.354 |
| 16 |
251028_at |
AT5G02230
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
0.551 |
0.330 |
| 17 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
0.528 |
0.408 |
| 18 |
247600_at |
AT5G60890
|
ATMYB34, ATR1, ALTERED TRYPTOPHAN REGULATION 1, MYB34, myb domain protein 34 |
0.526 |
-0.051 |
| 19 |
249205_at |
AT5G42600
|
MRN1, marneral synthase 1 |
0.522 |
-0.022 |
| 20 |
254056_at |
AT4G25250
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.519 |
0.082 |
| 21 |
259264_at |
AT3G01260
|
[Galactose mutarotase-like superfamily protein] |
0.500 |
-0.665 |
| 22 |
255437_at |
AT4G03060
|
AOP2, alkenyl hydroxalkyl producing 2 |
0.498 |
0.245 |
| 23 |
262608_at |
AT1G14120
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.493 |
0.048 |
| 24 |
253657_at |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
0.493 |
0.231 |
| 25 |
255604_at |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
0.491 |
0.479 |
| 26 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.491 |
0.382 |
| 27 |
258629_at |
AT3G02850
|
SKOR, STELAR K+ outward rectifier |
0.491 |
-0.281 |
| 28 |
249850_at |
AT5G23240
|
DJC76, DNA J protein C76 |
0.489 |
0.066 |
| 29 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.488 |
-0.035 |
| 30 |
264497_at |
AT1G30840
|
ATPUP4, purine permease 4, PUP4, purine permease 4 |
0.486 |
-0.370 |
| 31 |
262811_at |
AT1G11700
|
unknown |
0.484 |
0.452 |
| 32 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.471 |
0.158 |
| 33 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
0.469 |
0.412 |
| 34 |
256875_at |
AT3G26330
|
CYP71B37, cytochrome P450, family 71, subfamily B, polypeptide 37 |
0.465 |
-0.070 |
| 35 |
254305_at |
AT4G22200
|
AKT2, AKT2/3, potassium transport 2/3, AKT3, KT2/3, potassium transport 2/3 |
0.460 |
0.259 |
| 36 |
267101_at |
AT2G41480
|
AtPRX25, PRX25, peroxidase 25 |
0.456 |
0.219 |
| 37 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.447 |
0.301 |
| 38 |
245501_at |
AT4G15620
|
[Uncharacterised protein family (UPF0497)] |
0.445 |
0.147 |
| 39 |
258037_at |
AT3G21230
|
4CL5, 4-coumarate:CoA ligase 5 |
0.440 |
0.140 |
| 40 |
253411_at |
AT4G32980
|
ATH1, homeobox gene 1 |
0.439 |
0.321 |
| 41 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.438 |
0.528 |
| 42 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
0.436 |
-0.006 |
| 43 |
248372_at |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
0.433 |
0.296 |
| 44 |
266222_at |
AT2G28780
|
unknown |
0.429 |
-0.391 |
| 45 |
248353_at |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
0.425 |
0.233 |
| 46 |
253101_at |
AT4G37430
|
CYP81F1, CYTOCHROME P450 MONOOXYGENASE 81F1, CYP91A2, cytochrome P450, family 91, subfamily A, polypeptide 2 |
0.424 |
0.141 |
| 47 |
245304_at |
AT4G15630
|
[Uncharacterised protein family (UPF0497)] |
0.413 |
0.033 |
| 48 |
254907_at |
AT4G11190
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.408 |
0.102 |
| 49 |
255622_at |
AT4G01070
|
GT72B1, UGT72B1, UDP-GLUCOSE-DEPENDENT GLUCOSYLTRANSFERASE 72 B1 |
0.408 |
0.283 |
| 50 |
252534_at |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
0.404 |
0.050 |
| 51 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.403 |
0.407 |
| 52 |
252888_at |
AT4G39210
|
APL3 |
0.403 |
0.522 |
| 53 |
266828_at |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
0.402 |
-0.047 |
| 54 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.402 |
0.291 |
| 55 |
250937_at |
AT5G03230
|
unknown |
0.401 |
0.259 |
| 56 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.397 |
0.470 |
| 57 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.396 |
0.368 |
| 58 |
253822_at |
AT4G28410
|
RSA1, Root System Architecture 1 |
0.393 |
-0.112 |
| 59 |
266820_at |
AT2G44940
|
[Integrase-type DNA-binding superfamily protein] |
0.386 |
-0.101 |
| 60 |
266419_at |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
0.386 |
-0.012 |
| 61 |
248185_at |
AT5G54060
|
UF3GT, UDP-glucose:flavonoid 3-o-glucosyltransferase, UGT79B1, UDP-glucosyl transferase 79B1 |
0.382 |
0.181 |
| 62 |
245346_at |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
0.382 |
0.420 |
| 63 |
258078_at |
AT3G25870
|
unknown |
0.381 |
0.321 |
| 64 |
257985_at |
AT3G20810
|
JMJ30, Jumonji C domain-containing protein 30, JMJD5, jumonji domain containing 5 |
0.379 |
-0.087 |
| 65 |
246216_at |
AT4G36380
|
ROT3, ROTUNDIFOLIA 3 |
0.379 |
0.170 |
| 66 |
250216_at |
AT5G14090
|
AtLAZY1, LAZY1, LAZY 1 |
0.377 |
0.112 |
| 67 |
253619_at |
AT4G30460
|
[glycine-rich protein] |
0.375 |
-0.201 |
| 68 |
263452_at |
AT2G22190
|
TPPE, trehalose-6-phosphate phosphatase E |
0.373 |
0.139 |
| 69 |
252661_at |
AT3G44450
|
unknown |
0.372 |
0.158 |
| 70 |
261037_at |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
0.372 |
0.398 |
| 71 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.368 |
0.325 |
| 72 |
266578_at |
AT2G23910
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.367 |
-0.058 |
| 73 |
254631_at |
AT4G18610
|
LSH9, LIGHT SENSITIVE HYPOCOTYLS 9 |
0.367 |
0.048 |
| 74 |
259036_at |
AT3G09220
|
LAC7, laccase 7 |
0.363 |
0.120 |
| 75 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.361 |
0.175 |
| 76 |
247213_at |
AT5G64900
|
ATPEP1, ARABIDOPSIS THALIANA PEPTIDE 1, PEP1, PEPTIDE 1, PROPEP1, precursor of peptide 1 |
0.360 |
0.103 |
| 77 |
245901_at |
AT5G11060
|
KNAT4, KNOTTED1-like homeobox gene 4 |
0.358 |
0.342 |
| 78 |
261504_at |
AT1G71692
|
AGL12, AGAMOUS-like 12, XAL1, XAANTAL1 |
0.358 |
-0.170 |
| 79 |
261335_at |
AT1G44800
|
SIAR1, Siliques Are Red 1, UMAMIT18, Usually multiple acids move in and out Transporters 18 |
0.356 |
0.204 |
| 80 |
253309_at |
AT4G33790
|
CER4, ECERIFERUM 4, FAR3, FATTY ACID REDUCTASE 3, G7 |
0.355 |
-0.161 |
| 81 |
246522_at |
AT5G15830
|
AtbZIP3, basic leucine-zipper 3, bZIP3, basic leucine-zipper 3 |
0.354 |
0.101 |
| 82 |
253697_at |
AT4G29700
|
[Alkaline-phosphatase-like family protein] |
0.353 |
-0.041 |
| 83 |
249046_at |
AT5G44400
|
[FAD-binding Berberine family protein] |
0.352 |
0.126 |
| 84 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.352 |
0.034 |
| 85 |
255575_at |
AT4G01430
|
UMAMIT29, Usually multiple acids move in and out Transporters 29 |
0.351 |
0.106 |
| 86 |
255621_at |
AT4G01390
|
[TRAF-like family protein] |
0.350 |
0.030 |
| 87 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.350 |
0.307 |
| 88 |
260262_at |
AT1G68470
|
[Exostosin family protein] |
0.349 |
0.100 |
| 89 |
253957_at |
AT4G26320
|
AGP13, arabinogalactan protein 13 |
0.345 |
-0.024 |
| 90 |
259010_at |
AT3G07340
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.345 |
0.276 |
| 91 |
266299_at |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
0.345 |
0.168 |
| 92 |
249848_at |
AT5G23220
|
NIC3, nicotinamidase 3 |
0.343 |
-0.066 |
| 93 |
265053_at |
AT1G52000
|
[Mannose-binding lectin superfamily protein] |
0.343 |
0.109 |
| 94 |
248467_at |
AT5G50800
|
AtSWEET13, SWEET13 |
0.341 |
0.274 |
| 95 |
265962_at |
AT2G37460
|
UMAMIT12, Usually multiple acids move in and out Transporters 12 |
0.339 |
0.123 |
| 96 |
263913_at |
AT2G36570
|
PXC1, PXY/TDR-correlated 1 |
0.339 |
0.098 |
| 97 |
249567_at |
AT5G38020
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.336 |
-0.030 |
| 98 |
249203_at |
AT5G42590
|
CYP71A16, cytochrome P450, family 71, subfamily A, polypeptide 16, MRO, marneral oxidase |
0.336 |
0.083 |
| 99 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.332 |
0.441 |
| 100 |
261921_at |
AT1G65900
|
unknown |
0.330 |
0.045 |
| 101 |
253344_at |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.329 |
0.192 |
| 102 |
249047_at |
AT5G44410
|
[FAD-binding Berberine family protein] |
0.326 |
0.178 |
| 103 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.325 |
0.389 |
| 104 |
254240_at |
AT4G23496
|
SP1L5, SPIRAL1-like5 |
0.324 |
-0.032 |
| 105 |
256774_at |
AT3G13760
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.323 |
0.056 |
| 106 |
264148_at |
AT1G02220
|
ANAC003, NAC domain containing protein 3, NAC003, NAC domain containing protein 3 |
0.323 |
0.119 |
| 107 |
253525_at |
AT4G31330
|
unknown |
0.322 |
0.311 |
| 108 |
247215_at |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
0.321 |
-0.039 |
| 109 |
249775_at |
AT5G24160
|
SQE6, squalene monoxygenase 6 |
0.320 |
-0.012 |
| 110 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.319 |
0.273 |
| 111 |
255488_at |
AT4G02630
|
[Protein kinase superfamily protein] |
0.319 |
0.270 |
| 112 |
263232_at |
AT1G05700
|
[Leucine-rich repeat transmembrane protein kinase protein] |
0.317 |
0.198 |
| 113 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.316 |
0.185 |
| 114 |
249800_at |
AT5G23660
|
AtSWEET12, MTN3, homolog of Medicago truncatula MTN3, SWEET12 |
0.315 |
0.257 |
| 115 |
245253_at |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
0.313 |
0.044 |
| 116 |
264091_at |
AT1G79110
|
BRG2, BOI-related gene 2 |
0.313 |
0.361 |
| 117 |
260060_at |
AT1G73680
|
ALPHA DOX2, alpha dioxygenase, alpha-DOX2, alpha-dioxygenase 2 |
0.311 |
0.203 |
| 118 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.310 |
0.375 |
| 119 |
254543_at |
AT4G19810
|
ChiC, class V chitinase |
0.309 |
0.179 |
| 120 |
259348_at |
AT3G03770
|
[Leucine-rich repeat protein kinase family protein] |
0.307 |
0.289 |
| 121 |
261956_at |
AT1G64590
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.305 |
-0.208 |
| 122 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
0.305 |
0.183 |
| 123 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.305 |
0.092 |
| 124 |
257300_at |
AT3G28080
|
UMAMIT47, Usually multiple acids move in and out Transporters 47 |
0.304 |
0.278 |
| 125 |
248979_at |
AT5G45080
|
AtPP2-A6, phloem protein 2-A6, PP2-A6, phloem protein 2-A6 |
0.303 |
-0.069 |
| 126 |
259854_at |
AT1G72200
|
[RING/U-box superfamily protein] |
0.303 |
-0.016 |
| 127 |
248890_at |
AT5G46270
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
0.302 |
0.270 |
| 128 |
252921_at |
AT4G39030
|
EDS5, ENHANCED DISEASE SUSCEPTIBILITY 5, SCORD3, susceptible to coronatine-deficient Pst DC3000 3, SID1, SALICYLIC ACID INDUCTION DEFICIENT 1 |
0.299 |
0.022 |
| 129 |
260551_at |
AT2G43510
|
ATTI1, trypsin inhibitor protein 1, TI1, trypsin inhibitor protein 1 |
0.299 |
0.249 |
| 130 |
246826_at |
AT5G26310
|
UGT72E3 |
0.299 |
-0.058 |
| 131 |
255648_at |
AT4G00910
|
[Aluminium activated malate transporter family protein] |
0.299 |
-0.098 |
| 132 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.297 |
-0.141 |
| 133 |
258437_at |
AT3G16560
|
[Protein phosphatase 2C family protein] |
0.297 |
0.150 |
| 134 |
251928_at |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.296 |
-0.053 |
| 135 |
267123_at |
AT2G23560
|
ATMES7, ARABIDOPSIS THALIANA METHYL ESTERASE 7, MES7, methyl esterase 7 |
0.296 |
0.085 |
| 136 |
259871_at |
AT1G76800
|
[Vacuolar iron transporter (VIT) family protein] |
0.295 |
0.302 |
| 137 |
256878_at |
AT3G26460
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.295 |
-0.022 |
| 138 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.295 |
0.398 |
| 139 |
260531_at |
AT2G47240
|
CER8, ECERIFERUM 8, LACS1, LONG-CHAIN ACYL-COA SYNTHASE 1 |
0.294 |
0.118 |
| 140 |
265305_at |
AT2G20340
|
AAS, aromatic aldehyde synthase, AtAAS |
0.294 |
0.090 |
| 141 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.294 |
0.398 |
| 142 |
267036_at |
AT2G38465
|
unknown |
0.293 |
0.268 |
| 143 |
264898_at |
AT1G23205
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.292 |
0.209 |
| 144 |
262137_at |
AT1G77920
|
TGA7, TGACG sequence-specific binding protein 7 |
0.291 |
0.244 |
| 145 |
262134_at |
AT1G77990
|
AST56, SULTR2;2, SULPHATE TRANSPORTER 2;2 |
0.291 |
0.236 |
| 146 |
252612_at |
AT3G45160
|
[Putative membrane lipoprotein] |
0.291 |
-0.178 |
| 147 |
263998_at |
AT2G22510
|
[hydroxyproline-rich glycoprotein family protein] |
0.290 |
0.285 |
| 148 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
0.289 |
0.406 |
| 149 |
258299_at |
AT3G23410
|
ATFAO3, ARABIDOPSIS FATTY ALCOHOL OXIDASE 3, FAO3, fatty alcohol oxidase 3 |
0.289 |
0.228 |
| 150 |
263549_at |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
0.288 |
-0.131 |
| 151 |
266996_at |
AT2G34490
|
CYP710A2, cytochrome P450, family 710, subfamily A, polypeptide 2 |
0.288 |
0.231 |
| 152 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
0.288 |
0.099 |
| 153 |
247038_at |
AT5G67160
|
EPS1, ENHANCED PSEUDOMONAS SUSCEPTIBILTY 1 |
0.287 |
0.151 |
| 154 |
247867_at |
AT5G57630
|
CIPK21, CBL-interacting protein kinase 21, SnRK3.4, SNF1-RELATED PROTEIN KINASE 3.4 |
0.286 |
0.047 |
| 155 |
267288_at |
AT2G23680
|
[Cold acclimation protein WCOR413 family] |
0.286 |
0.119 |
| 156 |
250582_at |
AT5G07580
|
[Integrase-type DNA-binding superfamily protein] |
0.285 |
0.091 |
| 157 |
254998_at |
AT4G09760
|
[Protein kinase superfamily protein] |
0.285 |
0.214 |
| 158 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.283 |
0.344 |
| 159 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.283 |
0.362 |
| 160 |
258851_at |
AT3G03190
|
ATGSTF11, glutathione S-transferase F11, ATGSTF6, ARABIDOPSIS GLUTATHIONE-S-TRANSFERASE 6, GSTF11, glutathione S-transferase F11 |
0.283 |
-0.104 |
| 161 |
247716_at |
AT5G59350
|
unknown |
0.282 |
0.119 |
| 162 |
262516_at |
AT1G17190
|
ATGSTU26, glutathione S-transferase tau 26, GSTU26, glutathione S-transferase tau 26 |
0.282 |
0.041 |
| 163 |
252827_at |
AT4G39950
|
CYP79B2, cytochrome P450, family 79, subfamily B, polypeptide 2 |
0.280 |
0.058 |
| 164 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.280 |
0.384 |
| 165 |
252488_at |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
0.279 |
0.242 |
| 166 |
245546_at |
AT4G15290
|
ATCSLB05, ATCSLB5, CELLULOSE SYNTHASE LIKE 5, CSLB05 |
0.279 |
-0.045 |
| 167 |
256626_at |
AT3G20015
|
[Eukaryotic aspartyl protease family protein] |
0.278 |
-0.071 |
| 168 |
248839_at |
AT5G46690
|
bHLH071, beta HLH protein 71 |
0.276 |
0.165 |
| 169 |
260771_at |
AT1G49160
|
WNK7 |
0.274 |
0.161 |
| 170 |
251785_at |
AT3G55130
|
ABCG19, ATP-binding cassette G19, ATWBC19, white-brown complex homolog 19, WBC19, white-brown complex homolog 19 |
0.273 |
0.176 |
| 171 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.272 |
0.181 |
| 172 |
247549_at |
AT5G61420
|
AtMYB28, HAG1, HIGH ALIPHATIC GLUCOSINOLATE 1, MYB28, myb domain protein 28, PMG1, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 1 |
0.271 |
0.082 |
| 173 |
262286_at |
AT1G68585
|
unknown |
0.271 |
0.244 |
| 174 |
246998_at |
AT5G67370
|
CGLD27, CONSERVED IN THE GREEN LINEAGE AND DIATOMS 27 |
0.271 |
0.272 |
| 175 |
253510_at |
AT4G31730
|
GDU1, glutamine dumper 1 |
0.270 |
0.139 |
| 176 |
247668_at |
AT5G60100
|
APRR3, pseudo-response regulator 3, PRR3, pseudo-response regulator 3 |
0.269 |
-0.048 |
| 177 |
265430_at |
AT2G20700
|
LLG2, LORELEI-LIKE-GPI ANCHORED PROTEIN 2 |
0.269 |
0.162 |
| 178 |
251480_at |
AT3G59710
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.268 |
0.235 |
| 179 |
265008_at |
AT1G61560
|
ATMLO6, MILDEW RESISTANCE LOCUS O 6, MLO6, MILDEW RESISTANCE LOCUS O 6 |
0.268 |
-0.109 |
| 180 |
246682_at |
AT5G33290
|
XGD1, xylogalacturonan deficient 1 |
0.267 |
0.270 |
| 181 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.266 |
0.338 |
| 182 |
265122_at |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
0.264 |
0.258 |
| 183 |
250438_at |
AT5G10580
|
unknown |
0.263 |
0.012 |
| 184 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.263 |
0.320 |
| 185 |
248395_at |
AT5G52120
|
AtPP2-A14, phloem protein 2-A14, PP2-A14, phloem protein 2-A14 |
0.262 |
-0.070 |
| 186 |
256168_at |
AT1G51805
|
[Leucine-rich repeat protein kinase family protein] |
0.262 |
0.100 |
| 187 |
266275_at |
AT2G29370
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.261 |
0.281 |
| 188 |
246900_at |
AT5G25620
|
AtYUC6, YUC6, YUCCA6 |
0.261 |
0.066 |
| 189 |
257825_at |
AT3G26700
|
[Protein kinase superfamily protein] |
0.261 |
0.035 |
| 190 |
265894_at |
AT2G15050
|
LTP, lipid transfer protein, LTP7, lipid transfer protein 7 |
0.260 |
0.155 |
| 191 |
267240_at |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.260 |
0.259 |
| 192 |
262533_at |
AT1G17090
|
unknown |
0.259 |
0.059 |
| 193 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.258 |
0.292 |
| 194 |
249768_at |
AT5G24100
|
[Leucine-rich repeat protein kinase family protein] |
0.257 |
0.065 |
| 195 |
263715_at |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
0.257 |
0.230 |
| 196 |
247812_at |
AT5G58390
|
[Peroxidase superfamily protein] |
0.256 |
0.126 |
| 197 |
255578_at |
AT4G01450
|
UMAMIT30, Usually multiple acids move in and out Transporters 30 |
0.256 |
-0.031 |
| 198 |
262162_at |
AT1G78020
|
unknown |
0.255 |
0.195 |
| 199 |
259143_at |
AT3G10190
|
[Calcium-binding EF-hand family protein] |
0.255 |
0.055 |
| 200 |
247685_at |
AT5G59680
|
[Leucine-rich repeat protein kinase family protein] |
0.255 |
-0.098 |
| 201 |
262793_at |
AT1G13110
|
CYP71B7, cytochrome P450, family 71 subfamily B, polypeptide 7 |
0.254 |
0.087 |
| 202 |
261713_at |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
0.254 |
0.020 |
| 203 |
253088_at |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
0.253 |
0.023 |
| 204 |
254693_at |
AT4G17880
|
MYC4 |
0.253 |
0.074 |
| 205 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.252 |
0.085 |
| 206 |
262171_at |
AT1G74950
|
JAZ2, JASMONATE-ZIM-DOMAIN PROTEIN 2, TIFY10B |
0.252 |
0.091 |
| 207 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
0.252 |
-0.562 |
| 208 |
255438_at |
AT4G03070
|
AOP, AOP1, AOP1.1 |
0.252 |
0.025 |
| 209 |
264186_at |
AT1G54570
|
PES1, phytyl ester synthase 1 |
0.252 |
0.158 |
| 210 |
264652_at |
AT1G08920
|
ESL1, ERD (early response to dehydration) six-like 1 |
0.251 |
-0.049 |
| 211 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.251 |
0.157 |
| 212 |
251335_at |
AT3G61400
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.251 |
-0.090 |
| 213 |
250598_at |
AT5G07690
|
ATMYB29, myb domain protein 29, MYB29, myb domain protein 29, PMG2, PRODUCTION OF METHIONINE-DERIVED GLUCOSINOLATE 2 |
0.250 |
-0.016 |
| 214 |
267425_at |
AT2G34810
|
[FAD-binding Berberine family protein] |
0.250 |
0.101 |
| 215 |
252485_at |
AT3G46530
|
RPP13, RECOGNITION OF PERONOSPORA PARASITICA 13 |
0.250 |
0.206 |
| 216 |
246781_at |
AT5G27350
|
SFP1 |
0.249 |
0.170 |
| 217 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
0.248 |
-0.148 |
| 218 |
266611_at |
AT2G14960
|
GH3.1 |
-1.085 |
-0.476 |
| 219 |
251565_at |
AT3G58190
|
ASL16, ASYMMETRIC LEAVES 2-LIKE 16, LBD29, lateral organ boundaries-domain 29 |
-1.075 |
-0.230 |
| 220 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.965 |
-0.528 |
| 221 |
266974_at |
AT2G39370
|
MAKR4, MEMBRANE-ASSOCIATED KINASE REGULATOR 4 |
-0.962 |
-0.306 |
| 222 |
245140_at |
AT2G45420
|
LBD18, LOB domain-containing protein 18 |
-0.927 |
-0.375 |
| 223 |
250509_at |
AT5G09970
|
CYP78A7, cytochrome P450, family 78, subfamily A, polypeptide 7 |
-0.926 |
-0.383 |
| 224 |
261766_at |
AT1G15580
|
ATAUX2-27, AUX2-27, AUXIN-INDUCIBLE 2-27, IAA5, indole-3-acetic acid inducible 5 |
-0.926 |
-0.576 |
| 225 |
251436_at |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-0.909 |
-0.379 |
| 226 |
265806_at |
AT2G18010
|
SAUR10, SMALL AUXIN UPREGULATED RNA 10 |
-0.877 |
-0.462 |
| 227 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.874 |
-0.614 |
| 228 |
250709_at |
AT5G06080
|
LBD33, LOB domain-containing protein 33 |
-0.867 |
-0.307 |
| 229 |
247878_at |
AT5G57760
|
unknown |
-0.863 |
-0.577 |
| 230 |
247869_at |
AT5G57520
|
ATZFP2, ZINC FINGER PROTEIN 2, ZFP2, zinc finger protein 2 |
-0.856 |
-0.412 |
| 231 |
266737_at |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
-0.856 |
-0.385 |
| 232 |
253179_at |
AT4G35200
|
unknown |
-0.850 |
-0.458 |
| 233 |
250091_at |
AT5G17340
|
[Putative membrane lipoprotein] |
-0.819 |
-0.333 |
| 234 |
248208_at |
AT5G53980
|
ATHB52, homeobox protein 52, HB52, homeobox protein 52 |
-0.817 |
-0.535 |
| 235 |
256010_at |
AT1G19220
|
ARF11, AUXIN RESPONSE FACTOR11, ARF19, auxin response factor 19, IAA22, indole-3-acetic acid inducible 22 |
-0.808 |
-0.204 |
| 236 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
-0.801 |
-0.364 |
| 237 |
254323_at |
AT4G22620
|
SAUR34, SMALL AUXIN UPREGULATED RNA 34 |
-0.799 |
-0.277 |
| 238 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.791 |
-0.411 |
| 239 |
246000_at |
AT5G20820
|
SAUR76, SMALL AUXIN UPREGULATED RNA 76 |
-0.768 |
-0.188 |
| 240 |
256743_at |
AT3G29370
|
P1R3, P1R3 |
-0.766 |
-0.454 |
| 241 |
252103_at |
AT3G51410
|
unknown |
-0.765 |
-0.403 |
| 242 |
248164_at |
AT5G54490
|
PBP1, pinoid-binding protein 1 |
-0.753 |
-0.455 |
| 243 |
246798_at |
AT5G26930
|
GATA23, GATA transcription factor 23 |
-0.746 |
-0.432 |
| 244 |
247474_at |
AT5G62280
|
unknown |
-0.743 |
-0.247 |
| 245 |
248509_at |
AT5G50335
|
unknown |
-0.738 |
-0.452 |
| 246 |
253155_at |
AT4G35720
|
unknown |
-0.716 |
-0.660 |
| 247 |
247951_at |
AT5G57240
|
ORP4C, OSBP(oxysterol binding protein)-related protein 4C |
-0.709 |
-0.293 |
| 248 |
267250_at |
AT2G23060
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.705 |
-0.340 |
| 249 |
246992_at |
AT5G67430
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
-0.701 |
-0.133 |
| 250 |
254809_at |
AT4G12410
|
SAUR35, SMALL AUXIN UPREGULATED RNA 35 |
-0.694 |
-0.308 |
| 251 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.693 |
-0.176 |
| 252 |
260608_at |
AT2G43870
|
[Pectin lyase-like superfamily protein] |
-0.690 |
-0.329 |
| 253 |
257386_at |
AT2G42440
|
ASL15, ASYMMETRIC LEAVES 2-like 15, LBD17, LOB domain-containing protein 17 |
-0.688 |
-0.276 |
| 254 |
245971_at |
AT5G20730
|
ARF7, AUXIN RESPONSE FACTOR 7, BIP, BIPOSTO, IAA21, indole-3-acetic acid inducible 21, IAA23, indole-3-acetic acid inducible 23, IAA25, indole-3-acetic acid inducible 25, MSG1, MASSUGU 1, NPH4, NON-PHOTOTROPHIC HYPOCOTYL, TIR5, TRANSPORT INHIBITOR RESPONSE 5 |
-0.686 |
0.039 |
| 255 |
247023_at |
AT5G67060
|
HEC1, HECATE 1 |
-0.685 |
-0.151 |
| 256 |
259027_at |
AT3G09280
|
unknown |
-0.682 |
-0.251 |
| 257 |
262525_at |
AT1G17060
|
CHI2, CHIBI 2, CYP72C1, cytochrome p450 72c1, SHK1, SHRINK 1, SOB7, SUPPRESSOR OF PHYB-4 7 |
-0.681 |
-0.105 |
| 258 |
250327_at |
AT5G12050
|
unknown |
-0.681 |
-0.459 |
| 259 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-0.679 |
-0.284 |
| 260 |
267337_at |
AT2G39980
|
[HXXXD-type acyl-transferase family protein] |
-0.676 |
-0.129 |
| 261 |
253044_at |
AT4G37290
|
unknown |
-0.675 |
-0.323 |
| 262 |
256799_at |
AT3G18560
|
unknown |
-0.673 |
-0.437 |
| 263 |
252970_at |
AT4G38850
|
ATSAUR15, ARABIDOPSIS THALIANA SMALL AUXIN UPREGULATED 15, SAUR-AC1, ARABIDOPSIS COLUMBIA SAUR GENE 1, SAUR15, SMALL AUXIN UPREGULATED 15, SAUR_AC1, SMALL AUXIN UP RNA 1 FROM ARABIDOPSIS THALIANA ECOTYPE COLUMBIA |
-0.668 |
-0.370 |
| 264 |
251612_at |
AT3G57950
|
unknown |
-0.668 |
-0.229 |
| 265 |
259785_at |
AT1G29490
|
SAUR68, SMALL AUXIN UPREGULATED 68 |
-0.665 |
-0.382 |
| 266 |
245416_at |
AT4G17350
|
unknown |
-0.664 |
-0.273 |
| 267 |
259735_at |
AT1G64405
|
unknown |
-0.662 |
-0.249 |
| 268 |
259331_at |
AT3G03840
|
SAUR27, SMALL AUXIN UP RNA 27 |
-0.662 |
-0.383 |
| 269 |
246932_at |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.657 |
-0.180 |
| 270 |
260668_at |
AT1G19530
|
unknown |
-0.655 |
-0.068 |
| 271 |
258253_at |
AT3G26760
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.647 |
-0.324 |
| 272 |
253103_at |
AT4G36110
|
SAUR9, SMALL AUXIN UPREGULATED RNA 9 |
-0.645 |
-0.296 |
| 273 |
251293_at |
AT3G61930
|
unknown |
-0.629 |
-0.191 |
| 274 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.628 |
-0.406 |
| 275 |
253066_at |
AT4G37770
|
ACS8, 1-amino-cyclopropane-1-carboxylate synthase 8 |
-0.624 |
-0.358 |
| 276 |
263653_at |
AT1G04310
|
ERS2, ethylene response sensor 2 |
-0.620 |
-0.079 |
| 277 |
249065_at |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-0.617 |
-0.150 |
| 278 |
251377_at |
AT3G60650
|
unknown |
-0.610 |
-0.251 |
| 279 |
248717_at |
AT5G48175
|
unknown |
-0.607 |
-0.187 |
| 280 |
260957_at |
AT1G06080
|
ADS1, delta 9 desaturase 1, AtADS1 |
-0.607 |
-0.391 |
| 281 |
250155_at |
AT5G15160
|
BHLH134, BASIC HELIX-LOOP-HELIX PROTEIN 134, BNQ2, BANQUO 2 |
-0.605 |
-0.364 |
| 282 |
258878_at |
AT3G03170
|
unknown |
-0.604 |
-0.157 |
| 283 |
266364_at |
AT2G41230
|
ORS1, ORGAN SIZE RELATED 1 |
-0.604 |
-0.166 |
| 284 |
267357_at |
AT2G40000
|
ATHSPRO2, ARABIDOPSIS ORTHOLOG OF SUGAR BEET HS1 PRO-1 2, HSPRO2, ortholog of sugar beet HS1 PRO-1 2 |
-0.602 |
-0.223 |
| 285 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.601 |
-0.511 |
| 286 |
248282_at |
AT5G52900
|
MAKR6, MEMBRANE-ASSOCIATED KINASE REGULATOR 6 |
-0.597 |
0.009 |
| 287 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.597 |
-0.317 |
| 288 |
255695_at |
AT4G00080
|
UNE11, unfertilized embryo sac 11 |
-0.596 |
-0.573 |
| 289 |
253047_at |
AT4G37295
|
unknown |
-0.596 |
-0.156 |
| 290 |
262448_at |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.585 |
-0.214 |
| 291 |
249992_at |
AT5G18560
|
PUCHI |
-0.583 |
-0.288 |
| 292 |
247179_at |
AT5G65320
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.581 |
0.014 |
| 293 |
264788_at |
AT2G17880
|
DJC24, DNA J protein C24 |
-0.579 |
-0.273 |
| 294 |
254459_at |
AT4G21200
|
ATGA2OX8, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 8, GA2OX8, gibberellin 2-oxidase 8 |
-0.577 |
-0.295 |
| 295 |
266830_at |
AT2G22810
|
ACC4, 1-AMINOCYCLOPROPANE-1-CARBOXYLIC ACID SYNTHASE POLYPEPTIDE, ACS4, 1-aminocyclopropane-1-carboxylate synthase 4, ATACS4 |
-0.576 |
-0.337 |
| 296 |
258769_at |
AT3G10870
|
ATMES17, ARABIDOPSIS THALIANA METHYL ESTERASE 17, MES17, methyl esterase 17 |
-0.573 |
-0.145 |
| 297 |
245397_at |
AT4G14560
|
AXR5, AUXIN RESISTANT 5, IAA1, indole-3-acetic acid inducible 1 |
-0.570 |
-0.485 |
| 298 |
248623_at |
AT5G49170
|
unknown |
-0.567 |
-0.336 |
| 299 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
-0.562 |
-0.527 |
| 300 |
257264_at |
AT3G22060
|
[Receptor-like protein kinase-related family protein] |
-0.553 |
0.109 |
| 301 |
252765_at |
AT3G42800
|
unknown |
-0.549 |
-0.554 |
| 302 |
260915_at |
AT1G02660
|
[alpha/beta-Hydrolases superfamily protein] |
-0.525 |
-0.254 |
| 303 |
267372_at |
AT2G26290
|
ARSK1, root-specific kinase 1 |
-0.524 |
-0.247 |
| 304 |
253908_at |
AT4G27260
|
GH3.5, WES1 |
-0.524 |
-0.112 |
| 305 |
253207_at |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-0.522 |
-0.228 |
| 306 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.520 |
-0.142 |
| 307 |
256030_at |
AT1G34110
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.519 |
-0.231 |
| 308 |
251160_at |
AT3G63240
|
[DNAse I-like superfamily protein] |
-0.519 |
-0.204 |
| 309 |
257506_at |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.515 |
-0.495 |
| 310 |
258399_at |
AT3G15540
|
IAA19, indole-3-acetic acid inducible 19, MSG2, MASSUGU 2 |
-0.506 |
-0.541 |
| 311 |
263959_at |
AT2G36210
|
SAUR45, SMALL AUXIN UPREGULATED RNA 45 |
-0.505 |
0.058 |
| 312 |
252204_at |
AT3G50340
|
unknown |
-0.499 |
-0.167 |
| 313 |
254300_at |
AT4G22780
|
ACR7, ACT domain repeat 7 |
-0.497 |
-0.267 |
| 314 |
248456_at |
AT5G51380
|
[RNI-like superfamily protein] |
-0.497 |
-0.238 |
| 315 |
259787_at |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.497 |
-0.397 |
| 316 |
259332_at |
AT3G03830
|
SAUR28, SMALL AUXIN UP RNA 28 |
-0.496 |
-0.300 |
| 317 |
266832_at |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-0.494 |
-0.277 |
| 318 |
250907_at |
AT5G03670
|
TRM28, TON1 Recruiting Motif 28 |
-0.493 |
-0.172 |
| 319 |
264479_at |
AT1G77280
|
[Protein kinase protein with adenine nucleotide alpha hydrolases-like domain] |
-0.492 |
-0.189 |
| 320 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.492 |
-0.525 |
| 321 |
258091_at |
AT3G14560
|
unknown |
-0.492 |
-0.543 |
| 322 |
257049_at |
AT3G15250
|
unknown |
-0.491 |
-0.193 |
| 323 |
256981_at |
AT3G13380
|
BRL3, BRI1-like 3 |
-0.489 |
0.006 |
| 324 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.484 |
-0.449 |
| 325 |
249939_at |
AT5G22430
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.477 |
-0.200 |
| 326 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.476 |
-0.367 |
| 327 |
253910_at |
AT4G27290
|
[S-locus lectin protein kinase family protein] |
-0.474 |
-0.303 |
| 328 |
266711_at |
AT2G46740
|
AtGulLO5, GulLO5, L -gulono-1,4-lactone ( L -GulL) oxidase 5 |
-0.472 |
-0.097 |
| 329 |
245208_at |
AT5G12330
|
LRP1, LATERAL ROOT PRIMORDIUM 1 |
-0.468 |
0.141 |
| 330 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.467 |
-0.600 |
| 331 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
-0.463 |
-0.274 |
| 332 |
253791_at |
AT4G28640
|
IAA11, indole-3-acetic acid inducible 11 |
-0.460 |
-0.112 |
| 333 |
261717_at |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-0.456 |
-0.236 |
| 334 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
-0.455 |
0.005 |
| 335 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.454 |
-0.713 |
| 336 |
252279_at |
AT3G49700
|
ACS9, 1-aminocyclopropane-1-carboxylate synthase 9, AtACS9, ETO3, ETHYLENE OVERPRODUCING 3 |
-0.454 |
-0.217 |
| 337 |
245196_at |
AT1G67750
|
[Pectate lyase family protein] |
-0.454 |
0.052 |
| 338 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.453 |
-0.493 |
| 339 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.447 |
-0.269 |
| 340 |
253011_at |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-0.445 |
-0.192 |
| 341 |
257481_at |
AT1G08430
|
ALMT1, aluminum-activated malate transporter 1, ATALMT1, ARABIDOPSIS THALIANA ALUMINUM-ACTIVATED MALATE TRANSPORTER 1 |
-0.445 |
-0.090 |
| 342 |
264511_at |
AT1G09350
|
AtGolS3, galactinol synthase 3, GolS3, galactinol synthase 3 |
-0.444 |
-0.247 |
| 343 |
247352_at |
AT5G63650
|
SNRK2-5, SNRK2.5, SNF1-related protein kinase 2.5, SRK2H, SNF1-RELATED PROTEIN KINASE 2H |
-0.444 |
-0.205 |
| 344 |
263436_at |
AT2G28690
|
unknown |
-0.444 |
0.003 |
| 345 |
265856_at |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-0.444 |
-0.247 |
| 346 |
251751_at |
AT3G55720
|
unknown |
-0.443 |
-0.281 |
| 347 |
255177_at |
AT4G08040
|
ACS11, 1-aminocyclopropane-1-carboxylate synthase 11 |
-0.443 |
-0.181 |
| 348 |
261470_at |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
-0.442 |
-0.217 |
| 349 |
258468_at |
AT3G06070
|
unknown |
-0.442 |
-0.459 |
| 350 |
257026_at |
AT3G19200
|
unknown |
-0.439 |
-0.135 |
| 351 |
251065_at |
AT5G01870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.433 |
-0.047 |
| 352 |
260770_at |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.431 |
-0.262 |
| 353 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.431 |
-0.406 |
| 354 |
249895_at |
AT5G22500
|
FAR1, fatty acid reductase 1 |
-0.430 |
-0.311 |
| 355 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
-0.430 |
-0.309 |
| 356 |
261226_at |
AT1G20190
|
ATEXP11, ATEXPA11, expansin 11, ATHEXP ALPHA 1.14, EXP11, EXPANSIN 11, EXPA11, expansin 11 |
-0.429 |
-0.337 |
| 357 |
255250_at |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
-0.428 |
-0.271 |
| 358 |
249887_at |
AT5G22310
|
unknown |
-0.427 |
-0.294 |
| 359 |
255037_at |
AT4G09460
|
AtMYB6, myb domain protein 6, MYB6, myb domain protein 6 |
-0.422 |
-0.263 |
| 360 |
251010_at |
AT5G02550
|
unknown |
-0.422 |
-0.155 |
| 361 |
264527_at |
AT1G30760
|
[FAD-binding Berberine family protein] |
-0.422 |
0.379 |
| 362 |
248423_at |
AT5G51670
|
unknown |
-0.420 |
-0.319 |
| 363 |
259445_at |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
-0.417 |
-0.350 |
| 364 |
263207_at |
AT1G10550
|
XET, XYLOGLUCAN:XYLOGLUCOSYL TRANSFERASE 33, XTH33, xyloglucan:xyloglucosyl transferase 33 |
-0.417 |
-0.328 |
| 365 |
245141_at |
AT2G45400
|
BEN1, BRI1-5 ENHANCED 1 |
-0.417 |
-0.222 |
| 366 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.417 |
-0.446 |
| 367 |
267614_at |
AT2G26710
|
BAS1, PHYB ACTIVATION TAGGED SUPPRESSOR 1, CYP72B1, CYP734A1 |
-0.417 |
-0.342 |
| 368 |
250012_x_at |
AT5G18060
|
SAUR23, SMALL AUXIN UP RNA 23 |
-0.415 |
-0.356 |
| 369 |
260376_at |
AT1G74110
|
CYP78A10, cytochrome P450, family 78, subfamily A, polypeptide 10 |
-0.410 |
-0.162 |
| 370 |
253618_at |
AT4G30420
|
UMAMIT34, Usually multiple acids move in and out Transporters 34 |
-0.408 |
-0.199 |
| 371 |
256583_at |
AT3G28850
|
[Glutaredoxin family protein] |
-0.407 |
-0.252 |
| 372 |
265345_at |
AT2G22680
|
WAVH1, WAV3 homolog 1 |
-0.407 |
-0.051 |
| 373 |
260492_at |
AT2G41850
|
ADPG2, ARABIDOPSIS DEHISCENCE ZONE POLYGALACTURONASE 2, PGAZAT, polygalacturonase abscission zone A. thaliana |
-0.406 |
-0.024 |
| 374 |
248770_at |
AT5G47740
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.406 |
-0.245 |
| 375 |
261917_at |
AT1G65920
|
[Regulator of chromosome condensation (RCC1) family with FYVE zinc finger domain] |
-0.403 |
-0.204 |
| 376 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.402 |
-0.212 |
| 377 |
262001_at |
AT1G33790
|
[jacalin lectin family protein] |
-0.400 |
0.029 |
| 378 |
261777_at |
AT1G76210
|
unknown |
-0.400 |
-0.076 |
| 379 |
264467_at |
AT1G10140
|
[Uncharacterised conserved protein UCP031279] |
-0.399 |
-0.023 |
| 380 |
260841_at |
AT1G29195
|
unknown |
-0.398 |
-0.117 |
| 381 |
250746_at |
AT5G05880
|
[UDP-Glycosyltransferase superfamily protein] |
-0.395 |
-0.138 |
| 382 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.395 |
-0.476 |
| 383 |
250007_at |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-0.393 |
-0.000 |
| 384 |
250200_at |
AT5G14130
|
[Peroxidase superfamily protein] |
-0.392 |
-0.139 |
| 385 |
247991_at |
AT5G56320
|
ATEXP14, ATEXPA14, expansin A14, ATHEXP ALPHA 1.5, EXP14, EXPANSIN 14, EXPA14, expansin A14 |
-0.392 |
-0.241 |
| 386 |
255413_at |
AT4G03140
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.391 |
-0.208 |
| 387 |
258764_at |
AT3G10720
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.391 |
-0.196 |
| 388 |
259784_at |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.391 |
-0.403 |
| 389 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.390 |
-0.442 |
| 390 |
251434_at |
AT3G59850
|
[Pectin lyase-like superfamily protein] |
-0.389 |
-0.044 |
| 391 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
-0.388 |
-0.343 |
| 392 |
258516_at |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
-0.387 |
-0.160 |
| 393 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.387 |
-0.247 |
| 394 |
253660_at |
AT4G30140
|
CDEF1, CUTICLE DESTRUCTING FACTOR 1 |
-0.385 |
0.153 |
| 395 |
266507_at |
AT2G47860
|
SETH6 |
-0.383 |
-0.156 |
| 396 |
258718_at |
AT3G09760
|
[RING/U-box superfamily protein] |
-0.383 |
-0.176 |
| 397 |
260483_at |
AT1G11000
|
ATMLO4, MILDEW RESISTANCE LOCUS O 4, MLO4, MILDEW RESISTANCE LOCUS O 4 |
-0.382 |
-0.283 |
| 398 |
245277_at |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-0.381 |
-0.027 |
| 399 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.381 |
-0.750 |
| 400 |
265144_at |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
-0.380 |
-0.231 |
| 401 |
248910_at |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-0.380 |
-0.306 |
| 402 |
258388_at |
AT3G15370
|
ATEXP12, ATEXPA12, expansin 12, ATHEXP ALPHA 1.24, EXP12, EXPANSIN 12, EXPA12, expansin 12 |
-0.380 |
-0.193 |
| 403 |
248118_at |
AT5G55050
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-0.380 |
-0.121 |
| 404 |
266307_at |
AT2G27000
|
CYP705A8, cytochrome P450, family 705, subfamily A, polypeptide 8 |
-0.380 |
-0.362 |
| 405 |
245056_at |
AT2G26480
|
UGT76D1, UDP-glucosyl transferase 76D1 |
-0.377 |
-0.171 |
| 406 |
251144_at |
AT5G01210
|
[HXXXD-type acyl-transferase family protein] |
-0.374 |
-0.200 |
| 407 |
266368_at |
AT2G41380
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.374 |
-0.254 |
| 408 |
250145_at |
AT5G14690
|
unknown |
-0.374 |
-0.078 |
| 409 |
255528_at |
AT4G02090
|
unknown |
-0.373 |
-0.202 |
| 410 |
259120_at |
AT3G02240
|
GLV4, GOLVEN 4, RGF7, root meristem growth factor 7 |
-0.371 |
-0.141 |
| 411 |
247003_at |
AT5G67550
|
unknown |
-0.371 |
-0.104 |
| 412 |
248371_at |
AT5G51810
|
AT2353, ATGA20OX2, GA20OX2, gibberellin 20 oxidase 2 |
-0.371 |
-0.219 |
| 413 |
248564_at |
AT5G49700
|
AHL17, AT-hook motif nuclear localized protein 17 |
-0.371 |
0.071 |
| 414 |
248253_at |
AT5G53290
|
CRF3, cytokinin response factor 3 |
-0.370 |
-0.183 |
| 415 |
257373_at |
AT2G43140
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.370 |
-0.120 |
| 416 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-0.368 |
-0.607 |
| 417 |
256190_at |
AT1G30100
|
ATNCED5, NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 5, NCED5, nine-cis-epoxycarotenoid dioxygenase 5 |
-0.368 |
-0.136 |
| 418 |
257643_at |
AT3G25730
|
EDF3, ethylene response DNA binding factor 3 |
-0.368 |
-0.045 |
| 419 |
256714_at |
AT2G34080
|
[Cysteine proteinases superfamily protein] |
-0.367 |
0.219 |
| 420 |
266908_at |
AT2G34650
|
ABR, ABRUPTUS, PID, PINOID |
-0.367 |
-0.066 |
| 421 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.366 |
-0.201 |
| 422 |
249087_at |
AT5G44210
|
ATERF-9, ERF DOMAIN PROTEIN- 9, ATERF9, ERF DOMAIN PROTEIN 9, ERF9, erf domain protein 9 |
-0.364 |
-0.137 |
| 423 |
257927_at |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
-0.364 |
-0.169 |
| 424 |
259018_at |
AT3G07390
|
AIR12, Auxin-Induced in Root cultures 12 |
-0.363 |
-0.090 |
| 425 |
259268_at |
AT3G01070
|
AtENODL16, ENODL16, early nodulin-like protein 16 |
-0.360 |
-0.189 |
| 426 |
263360_at |
AT2G03830
|
GLV6, GOLVEN 6, RGF8, root meristem growth factor 8 |
-0.359 |
-0.050 |
| 427 |
262045_at |
AT1G80240
|
DGR1, DUF642 L-GalL responsive gene 1 |
-0.359 |
-0.477 |
| 428 |
250803_at |
AT5G04980
|
[DNAse I-like superfamily protein] |
-0.359 |
-0.298 |
| 429 |
263575_at |
AT2G17070
|
unknown |
-0.358 |
-0.217 |
| 430 |
258029_at |
AT3G27580
|
ATPK7, D6PKL3, D6 PROTEIN KINASE LIKE 3 |
-0.358 |
-0.101 |
| 431 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.358 |
-0.386 |
| 432 |
262971_at |
AT1G75640
|
[Leucine-rich receptor-like protein kinase family protein] |
-0.354 |
-0.019 |
| 433 |
248163_at |
AT5G54510
|
DFL1, DWARF IN LIGHT 1, GH3.6, Gretchen Hagen3.6 |
-0.350 |
-0.437 |