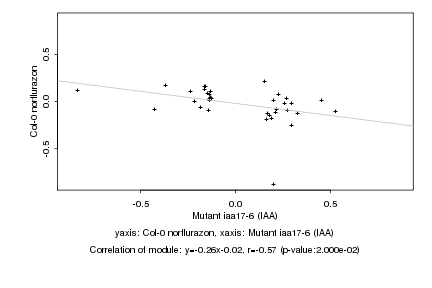
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant iaa17-6 (IAA) |
Log2 signal ratio
Col-0 norflurazon |
| 1 |
247718_at |
AT5G59310
|
LTP4, lipid transfer protein 4 |
0.523 |
-0.099 |
| 2 |
263851_at |
AT2G04460
|
unknown |
0.453 |
0.018 |
| 3 |
247717_at |
AT5G59320
|
LTP3, lipid transfer protein 3 |
0.323 |
-0.125 |
| 4 |
265846_at |
AT2G35770
|
scpl28, serine carboxypeptidase-like 28 |
0.295 |
-0.251 |
| 5 |
252533_at |
AT3G46110
|
[LOCATED IN: plasma membrane] |
0.294 |
-0.020 |
| 6 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.273 |
-0.088 |
| 7 |
252004_at |
AT3G52780
|
ATPAP20, PAP20 |
0.266 |
0.039 |
| 8 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.257 |
-0.011 |
| 9 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
0.224 |
0.078 |
| 10 |
249893_at |
AT5G22555
|
unknown |
0.215 |
-0.080 |
| 11 |
262236_at |
AT1G48330
|
unknown |
0.206 |
-0.110 |
| 12 |
250541_at |
AT5G09520
|
PELPK2, Pro-Glu-Leu|Ile|Val-Pro-Lys 2 |
0.196 |
-0.868 |
| 13 |
251621_at |
AT3G57700
|
[Protein kinase superfamily protein] |
0.196 |
0.014 |
| 14 |
247657_at |
AT5G59845
|
[Gibberellin-regulated family protein] |
0.186 |
-0.171 |
| 15 |
246395_at |
AT1G58170
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.175 |
-0.146 |
| 16 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
0.166 |
-0.126 |
| 17 |
265025_at |
AT1G24575
|
unknown |
0.160 |
-0.189 |
| 18 |
247610_at |
AT5G60630
|
unknown |
0.152 |
0.213 |
| 19 |
263664_at |
AT1G04250
|
AXR3, AUXIN RESISTANT 3, IAA17, indole-3-acetic acid inducible 17 |
-0.834 |
0.118 |
| 20 |
245449_at |
AT4G16870
|
[copia-like retrotransposon family, has a 0. P-value blast match to dbj|BAA78426.1| polyprotein (AtRE2-1) (Arabidopsis thaliana) (Ty1_Copia-element)] |
-0.428 |
-0.083 |
| 21 |
259252_at |
AT3G07610
|
IBM1, increase in bonsai methylation 1 |
-0.372 |
0.173 |
| 22 |
257444_at |
AT2G12550
|
NUB1, homolog of human NUB1 |
-0.239 |
0.107 |
| 23 |
259160_at |
AT3G05410
|
[Photosystem II reaction center PsbP family protein] |
-0.217 |
0.006 |
| 24 |
250979_at |
AT5G03090
|
unknown |
-0.186 |
-0.055 |
| 25 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
-0.167 |
0.163 |
| 26 |
262898_at |
AT1G59850
|
[ARM repeat superfamily protein] |
-0.164 |
0.128 |
| 27 |
263019_at |
AT1G23870
|
ATTPS9, trehalose-phosphatase/synthase 9, TPS9, TREHALOSE -6-PHOSPHATASE SYNTHASE S9, TPS9, trehalose-phosphatase/synthase 9 |
-0.158 |
0.165 |
| 28 |
263708_at |
AT1G09320
|
[agenet domain-containing protein] |
-0.148 |
0.091 |
| 29 |
255732_at |
AT1G25450
|
CER60, ECERIFERUM 60, KCS5, 3-ketoacyl-CoA synthase 5 |
-0.145 |
-0.094 |
| 30 |
250223_at |
AT5G14070
|
ROXY2 |
-0.139 |
0.078 |
| 31 |
255307_at |
AT4G04900
|
RIC10, ROP-interactive CRIB motif-containing protein 10 |
-0.139 |
0.014 |
| 32 |
251863_at |
AT3G54870
|
ARK1, ARMADILLO REPEAT-CONTAINING KINESIN 1, AtKINUc, Arabidopsis thaliana KINESIN Ungrouped clade, gene A, CAE1, CA-ROP2 ENHANCER 1, MRH2, MORPHOGENESIS OF ROOT HAIR 2 |
-0.138 |
0.034 |
| 33 |
260366_at |
AT1G70460
|
AtPERK13, proline-rich extensin-like receptor kinase 13, PERK13, proline-rich extensin-like receptor kinase 13, RHS10, root hair specific 10 |
-0.136 |
0.114 |
| 34 |
248429_at |
AT5G51770
|
[Protein kinase superfamily protein] |
-0.135 |
0.046 |
| 35 |
253794_at |
AT4G28720
|
YUC8, YUCCA 8 |
-0.129 |
0.040 |