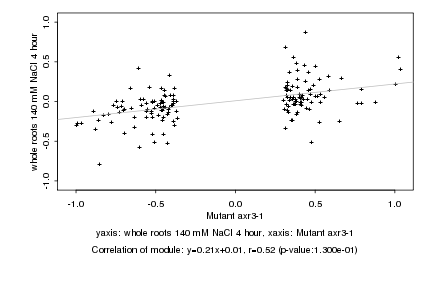
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant axr3-1 |
Log2 signal ratio
whole roots 140 mM NaCl 4 hour |
| 1 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
1.033 |
0.411 |
| 2 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
1.020 |
0.564 |
| 3 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.000 |
0.220 |
| 4 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.874 |
-0.003 |
| 5 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.790 |
0.160 |
| 6 |
263829_at |
AT2G40435
|
unknown |
0.786 |
-0.016 |
| 7 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.761 |
-0.013 |
| 8 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.665 |
0.292 |
| 9 |
262325_at |
AT1G64160
|
AtDIR5, DIR5, dirigent protein 5 |
0.652 |
-0.250 |
| 10 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.590 |
0.142 |
| 11 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.582 |
0.321 |
| 12 |
257089_at |
AT3G20520
|
GDPDL5, Glycerophosphodiester phosphodiesterase (GDPD) like 5, SVL3, SHV3-like 3 |
0.557 |
0.053 |
| 13 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.528 |
0.090 |
| 14 |
264469_at |
AT1G67100
|
LBD40, LOB domain-containing protein 40 |
0.528 |
-0.004 |
| 15 |
245173_at |
AT2G47520
|
AtERF71, Arabidopsis thaliana ethylene response factor 71, ERF71, ethylene response factor 71, HRE2, HYPOXIA RESPONSIVE ERF (ETHYLENE RESPONSE FACTOR) 2 |
0.527 |
-0.259 |
| 16 |
252888_at |
AT4G39210
|
APL3 |
0.522 |
0.280 |
| 17 |
252746_at |
AT3G43190
|
ATSUS4, ARABIDOPSIS THALIANA SUCROSE SYNTHASE 4, SUS4, sucrose synthase 4 |
0.513 |
0.066 |
| 18 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.498 |
0.448 |
| 19 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.496 |
0.068 |
| 20 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.487 |
0.212 |
| 21 |
261221_at |
AT1G19960
|
unknown |
0.476 |
-0.514 |
| 22 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
0.474 |
-0.002 |
| 23 |
250435_at |
AT5G10380
|
ATRING1, RING1 |
0.470 |
0.099 |
| 24 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.467 |
0.153 |
| 25 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.459 |
-0.099 |
| 26 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.456 |
0.371 |
| 27 |
256789_at |
AT3G13672
|
SINA2 |
0.455 |
0.139 |
| 28 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.449 |
0.036 |
| 29 |
245262_at |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
0.441 |
-0.078 |
| 30 |
255795_at |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
0.437 |
0.875 |
| 31 |
251858_at |
AT3G54820
|
PIP2;5, plasma membrane intrinsic protein 2;5, PIP2D, PLASMA MEMBRANE INTRINSIC PROTEIN 2D |
0.434 |
0.255 |
| 32 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.432 |
0.465 |
| 33 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.421 |
0.038 |
| 34 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.418 |
0.085 |
| 35 |
249215_at |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
0.413 |
0.051 |
| 36 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
0.413 |
-0.041 |
| 37 |
247447_at |
AT5G62730
|
[Major facilitator superfamily protein] |
0.412 |
-0.000 |
| 38 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
0.408 |
0.042 |
| 39 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.407 |
-0.012 |
| 40 |
254564_at |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
0.406 |
-0.028 |
| 41 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.398 |
0.089 |
| 42 |
266617_at |
AT2G29670
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.398 |
0.061 |
| 43 |
265766_at |
AT2G48080
|
[oxidoreductase, 2OG-Fe(II) oxygenase family protein] |
0.389 |
0.177 |
| 44 |
264403_at |
AT2G25150
|
[HXXXD-type acyl-transferase family protein] |
0.387 |
-0.130 |
| 45 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.385 |
0.284 |
| 46 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.384 |
0.396 |
| 47 |
250083_at |
AT5G17220
|
ATGSTF12, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE PHI 12, GST26, GLUTATHIONE S-TRANSFERASE 26, GSTF12, glutathione S-transferase phi 12, TT19, TRANSPARENT TESTA 19 |
0.382 |
-0.156 |
| 48 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.379 |
0.488 |
| 49 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.377 |
0.007 |
| 50 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
0.375 |
-0.033 |
| 51 |
248812_at |
AT5G47330
|
[alpha/beta-Hydrolases superfamily protein] |
0.375 |
0.009 |
| 52 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.373 |
0.034 |
| 53 |
249955_at |
AT5G18840
|
[Major facilitator superfamily protein] |
0.368 |
-0.028 |
| 54 |
259590_at |
AT1G28160
|
[Integrase-type DNA-binding superfamily protein] |
0.363 |
0.062 |
| 55 |
256244_at |
AT3G12520
|
SULTR4;2, sulfate transporter 4;2 |
0.362 |
-0.033 |
| 56 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.360 |
0.566 |
| 57 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.354 |
0.029 |
| 58 |
252316_at |
AT3G48700
|
ATCXE13, carboxyesterase 13, CXE13, carboxyesterase 13 |
0.353 |
0.192 |
| 59 |
262373_at |
AT1G73120
|
unknown |
0.352 |
-0.227 |
| 60 |
248961_at |
AT5G45650
|
[subtilase family protein] |
0.349 |
-0.229 |
| 61 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.344 |
0.058 |
| 62 |
257517_at |
AT3G16330
|
unknown |
0.342 |
0.141 |
| 63 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.338 |
0.019 |
| 64 |
262644_at |
AT1G62710
|
BETA-VPE, beta vacuolar processing enzyme, BETAVPE |
0.333 |
0.368 |
| 65 |
254242_at |
AT4G23200
|
CRK12, cysteine-rich RLK (RECEPTOR-like protein kinase) 12 |
0.332 |
-0.052 |
| 66 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
0.331 |
-0.136 |
| 67 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.326 |
0.157 |
| 68 |
248777_at |
AT5G47920
|
unknown |
0.326 |
0.202 |
| 69 |
256489_at |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.325 |
-0.102 |
| 70 |
251713_at |
AT3G56080
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
0.323 |
0.245 |
| 71 |
245528_at |
AT4G15530
|
PPDK, pyruvate orthophosphate dikinase |
0.322 |
0.059 |
| 72 |
260869_at |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN Δ9-DESATURASE6 |
0.321 |
0.147 |
| 73 |
266209_at |
AT2G27550
|
ATC, centroradialis |
0.320 |
0.076 |
| 74 |
263228_at |
AT1G30700
|
[FAD-binding Berberine family protein] |
0.318 |
0.144 |
| 75 |
265769_at |
AT2G48090
|
unknown |
0.318 |
0.168 |
| 76 |
252896_at |
AT4G39480
|
CYP96A9, cytochrome P450, family 96, subfamily A, polypeptide 9 |
0.316 |
-0.027 |
| 77 |
260335_at |
AT1G74000
|
SS3, strictosidine synthase 3 |
0.313 |
0.188 |
| 78 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.311 |
0.682 |
| 79 |
253525_at |
AT4G31330
|
unknown |
0.311 |
-0.331 |
| 80 |
259302_at |
AT3G05120
|
ATGID1A, GA INSENSITIVE DWARF1A, GID1A, GA INSENSITIVE DWARF1A |
0.307 |
-0.099 |
| 81 |
245624_at |
AT4G14090
|
[UDP-Glycosyltransferase superfamily protein] |
0.301 |
0.022 |
| 82 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-1.001 |
-0.291 |
| 83 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-0.991 |
-0.270 |
| 84 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-0.966 |
-0.268 |
| 85 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.891 |
-0.121 |
| 86 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.879 |
-0.348 |
| 87 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.862 |
-0.236 |
| 88 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.857 |
-0.781 |
| 89 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.829 |
-0.164 |
| 90 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.797 |
-0.156 |
| 91 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.782 |
-0.260 |
| 92 |
258746_at |
AT3G05950
|
[RmlC-like cupins superfamily protein] |
-0.767 |
-0.040 |
| 93 |
259783_at |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.750 |
0.008 |
| 94 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.741 |
-0.068 |
| 95 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.729 |
-0.136 |
| 96 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.721 |
-0.057 |
| 97 |
259773_at |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.713 |
0.002 |
| 98 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.704 |
-0.110 |
| 99 |
252296_at |
AT3G48970
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.702 |
-0.402 |
| 100 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.699 |
-0.100 |
| 101 |
253155_at |
AT4G35720
|
unknown |
-0.660 |
0.166 |
| 102 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.654 |
-0.084 |
| 103 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.638 |
-0.318 |
| 104 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.638 |
-0.195 |
| 105 |
255822_at |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.614 |
0.427 |
| 106 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-0.607 |
-0.576 |
| 107 |
263656_at |
AT1G04240
|
IAA3, indole-3-acetic acid inducible 3, SHY2, SHORT HYPOCOTYL 2 |
-0.600 |
0.026 |
| 108 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.590 |
-0.050 |
| 109 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.578 |
0.033 |
| 110 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.563 |
-0.114 |
| 111 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.562 |
-0.192 |
| 112 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.559 |
-0.019 |
| 113 |
252765_at |
AT3G42800
|
unknown |
-0.554 |
-0.091 |
| 114 |
258091_at |
AT3G14560
|
unknown |
-0.543 |
0.185 |
| 115 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.529 |
-0.148 |
| 116 |
251423_at |
AT3G60550
|
CYCP3;2, cyclin p3;2 |
-0.527 |
-0.120 |
| 117 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.526 |
-0.198 |
| 118 |
245336_at |
AT4G16515
|
CLEL 6, CLE-like 6, GLV1, GOLVEN 1, RGF6, root meristem growth factor 6 |
-0.525 |
0.002 |
| 119 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.522 |
0.000 |
| 120 |
264734_at |
AT1G62280
|
SLAH1, SLAC1 homologue 1 |
-0.521 |
-0.409 |
| 121 |
264157_at |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
-0.514 |
-0.511 |
| 122 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.513 |
-0.078 |
| 123 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.511 |
0.003 |
| 124 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.493 |
-0.040 |
| 125 |
264029_at |
AT2G03720
|
MRH6, morphogenesis of root hair 6 |
-0.487 |
-0.166 |
| 126 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.476 |
-0.065 |
| 127 |
260012_at |
AT1G67865
|
unknown |
-0.473 |
-0.014 |
| 128 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.469 |
0.013 |
| 129 |
254512_at |
AT4G20230
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.468 |
-0.111 |
| 130 |
247469_at |
AT5G62165
|
AGL42, AGAMOUS-like 42, FYF, FOREVER YOUNG FLOWER |
-0.464 |
0.176 |
| 131 |
258468_at |
AT3G06070
|
unknown |
-0.459 |
-0.003 |
| 132 |
263552_x_at |
AT2G24980
|
EXT6, extensin 6 |
-0.458 |
-0.102 |
| 133 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.458 |
-0.230 |
| 134 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.456 |
-0.010 |
| 135 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.455 |
-0.201 |
| 136 |
264342_at |
AT1G12080
|
[Vacuolar calcium-binding protein-related] |
-0.453 |
-0.404 |
| 137 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.453 |
-0.077 |
| 138 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.453 |
-0.053 |
| 139 |
263981_at |
AT2G42870
|
HLH1, HELIX-LOOP-HELIX 1, PAR1, PHY RAPIDLY REGULATED 1 |
-0.449 |
0.148 |
| 140 |
257701_at |
AT3G12710
|
[DNA glycosylase superfamily protein] |
-0.446 |
0.084 |
| 141 |
250778_at |
AT5G05500
|
MOP10 |
-0.445 |
-0.068 |
| 142 |
251575_at |
AT3G58120
|
ATBZIP61, BZIP61 |
-0.442 |
0.071 |
| 143 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.430 |
-0.156 |
| 144 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.428 |
-0.526 |
| 145 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.425 |
-0.135 |
| 146 |
258080_at |
AT3G25930
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.423 |
-0.090 |
| 147 |
246229_at |
AT4G37160
|
sks15, SKU5 similar 15 |
-0.417 |
-0.075 |
| 148 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.415 |
0.334 |
| 149 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.410 |
0.078 |
| 150 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.404 |
-0.046 |
| 151 |
260475_at |
AT1G11080
|
scpl31, serine carboxypeptidase-like 31 |
-0.396 |
-0.049 |
| 152 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.394 |
0.003 |
| 153 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.394 |
-0.021 |
| 154 |
256736_at |
AT3G29410
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
-0.394 |
-0.243 |
| 155 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.393 |
0.030 |
| 156 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.393 |
0.081 |
| 157 |
251195_at |
AT3G62930
|
[Thioredoxin superfamily protein] |
-0.388 |
-0.291 |
| 158 |
260527_at |
AT2G47270
|
UPB1, UPBEAT1 |
-0.386 |
0.170 |
| 159 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.376 |
0.011 |
| 160 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.375 |
-0.117 |
| 161 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.369 |
-0.208 |