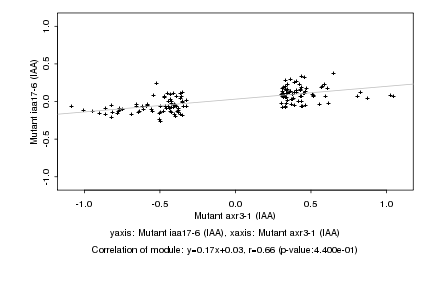
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant axr3-1 (IAA) |
Log2 signal ratio
Mutant iaa17-6 (IAA) |
| 1 |
252269_at |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
1.042 |
0.079 |
| 2 |
259329_at |
AT3G16360
|
AHP4, HPT phosphotransmitter 4 |
1.025 |
0.081 |
| 3 |
267389_at |
AT2G44460
|
BGLU28, beta glucosidase 28 |
0.873 |
0.048 |
| 4 |
258983_at |
AT3G08860
|
PYD4, PYRIMIDINE 4 |
0.822 |
0.120 |
| 5 |
248676_at |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
0.807 |
0.079 |
| 6 |
250142_at |
AT5G14650
|
[Pectin lyase-like superfamily protein] |
0.647 |
0.381 |
| 7 |
256319_at |
AT1G35910
|
TPPD, trehalose-6-phosphate phosphatase D |
0.611 |
-0.017 |
| 8 |
262357_at |
AT1G73040
|
[Mannose-binding lectin superfamily protein] |
0.604 |
0.186 |
| 9 |
257089_at |
AT3G20520
|
GDPDL5, Glycerophosphodiester phosphodiesterase (GDPD) like 5, SVL3, SHV3-like 3 |
0.590 |
0.077 |
| 10 |
260408_at |
AT1G69880
|
ATH8, thioredoxin H-type 8, TH8, thioredoxin H-type 8 |
0.587 |
0.234 |
| 11 |
262369_at |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
0.575 |
0.205 |
| 12 |
248227_at |
AT5G53820
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.567 |
0.191 |
| 13 |
251428_at |
AT3G60140
|
BGLU30, BETA GLUCOSIDASE 30, DIN2, DARK INDUCIBLE 2, SRG2, SENESCENCE-RELATED GENE 2 |
0.555 |
-0.028 |
| 14 |
246884_at |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
0.515 |
0.090 |
| 15 |
266561_at |
AT2G23960
|
[Class I glutamine amidotransferase-like superfamily protein] |
0.515 |
0.070 |
| 16 |
251898_at |
AT3G54340
|
AP3, APETALA 3, ATAP3 |
0.505 |
0.096 |
| 17 |
248777_at |
AT5G47920
|
unknown |
0.466 |
0.179 |
| 18 |
252415_at |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
0.462 |
-0.052 |
| 19 |
250702_at |
AT5G06730
|
[Peroxidase superfamily protein] |
0.458 |
0.142 |
| 20 |
255753_at |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
0.453 |
0.322 |
| 21 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
0.453 |
0.109 |
| 22 |
262154_at |
AT1G52700
|
[alpha/beta-Hydrolases superfamily protein] |
0.450 |
0.113 |
| 23 |
267384_at |
AT2G44370
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.440 |
-0.063 |
| 24 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
0.436 |
-0.059 |
| 25 |
256300_at |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
0.435 |
0.338 |
| 26 |
261221_at |
AT1G19960
|
unknown |
0.435 |
0.070 |
| 27 |
257624_at |
AT3G26220
|
CYP71B3, cytochrome P450, family 71, subfamily B, polypeptide 3 |
0.433 |
0.197 |
| 28 |
260623_at |
AT1G08090
|
ACH1, ATNRT2.1, NITRATE TRANSPORTER 2.1, ATNRT2:1, nitrate transporter 2:1, LIN1, LATERAL ROOT INITIATION 1, NRT2, NITRATE TRANSPORTER 2, NRT2.1, NITRATE TRANSPORTER 2.1, NRT2:1, nitrate transporter 2:1, NRT2;1AT |
0.432 |
0.013 |
| 29 |
259133_at |
AT3G05400
|
[Major facilitator superfamily protein] |
0.427 |
0.074 |
| 30 |
259789_at |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
0.426 |
0.157 |
| 31 |
258270_at |
AT3G15650
|
[alpha/beta-Hydrolases superfamily protein] |
0.426 |
0.173 |
| 32 |
245119_at |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
0.418 |
0.234 |
| 33 |
255958_at |
AT1G22150
|
SULTR1;3, sulfate transporter 1;3 |
0.415 |
0.007 |
| 34 |
249917_at |
AT5G22460
|
[alpha/beta-Hydrolases superfamily protein] |
0.404 |
0.273 |
| 35 |
253519_at |
AT4G31240
|
AtNRX2, NRX2, nucleoredoxin 2 |
0.400 |
0.153 |
| 36 |
266327_at |
AT2G46680
|
ATHB-7, homeobox 7, ATHB7, ARABIDOPSIS THALIANA HOMEOBOX 7, HB-7, homeobox 7 |
0.400 |
0.126 |
| 37 |
254153_at |
AT4G24450
|
ATGWD2, GWD3, PWD, phosphoglucan, water dikinase |
0.389 |
0.131 |
| 38 |
252629_at |
AT3G44970
|
[Cytochrome P450 superfamily protein] |
0.388 |
-0.042 |
| 39 |
246228_at |
AT4G36430
|
[Peroxidase superfamily protein] |
0.385 |
0.257 |
| 40 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.384 |
0.047 |
| 41 |
261899_at |
AT1G80820
|
ATCCR2, CCR2, cinnamoyl coa reductase |
0.376 |
0.096 |
| 42 |
258287_at |
AT3G15990
|
SULTR3;4, sulfate transporter 3;4 |
0.376 |
0.027 |
| 43 |
260890_at |
AT1G29090
|
[Cysteine proteinases superfamily protein] |
0.368 |
-0.039 |
| 44 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.368 |
0.032 |
| 45 |
259616_at |
AT1G47960
|
ATC/VIF1, CELL WALL / VACUOLAR INHIBITOR OF FRUCTOSIDASE 1, C/VIF1, cell wall / vacuolar inhibitor of fructosidase 1 |
0.364 |
0.137 |
| 46 |
255591_at |
AT4G01630
|
ATEXP17, EXPANSIN 17, ATEXPA17, expansin A17, ATHEXP ALPHA 1.13, EXPA17, expansin A17 |
0.363 |
0.300 |
| 47 |
254777_at |
AT4G12960
|
AtGILT, GILT, gamma-interferon-responsive lysosomal thiol reductase |
0.358 |
0.144 |
| 48 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.355 |
0.122 |
| 49 |
262905_at |
AT1G59730
|
ATH7, thioredoxin H-type 7, TH7, thioredoxin H-type 7 |
0.343 |
0.237 |
| 50 |
248855_at |
AT5G46590
|
anac096, NAC domain containing protein 96, NAC096, NAC domain containing protein 96 |
0.340 |
0.024 |
| 51 |
249752_at |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
0.339 |
0.117 |
| 52 |
263168_at |
AT1G03020
|
[Thioredoxin superfamily protein] |
0.339 |
0.107 |
| 53 |
265769_at |
AT2G48090
|
unknown |
0.338 |
0.160 |
| 54 |
260023_at |
AT1G30040
|
ATGA2OX2, gibberellin 2-oxidase, GA2OX2, gibberellin 2-oxidase, GA2OX2, GIBBERELLIN 2-OXIDASE 2 |
0.336 |
-0.014 |
| 55 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.335 |
0.155 |
| 56 |
260933_at |
AT1G02470
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
0.335 |
0.112 |
| 57 |
264314_at |
AT1G70420
|
unknown |
0.333 |
0.058 |
| 58 |
249101_at |
AT5G43580
|
UPI, UNUSUAL SERINE PROTEASE INHIBITOR |
0.330 |
0.211 |
| 59 |
253814_at |
AT4G28290
|
unknown |
0.330 |
-0.053 |
| 60 |
257625_at |
AT3G26230
|
CYP71B24, cytochrome P450, family 71, subfamily B, polypeptide 24 |
0.330 |
0.282 |
| 61 |
250798_at |
AT5G05340
|
PRX52, peroxidase 52 |
0.329 |
0.066 |
| 62 |
263380_at |
AT2G40200
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
0.329 |
-0.074 |
| 63 |
247208_at |
AT5G64870
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
0.327 |
0.171 |
| 64 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.321 |
0.085 |
| 65 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.317 |
0.196 |
| 66 |
262092_at |
AT1G56150
|
SAUR71, SMALL AUXIN UPREGULATED 71 |
0.315 |
0.114 |
| 67 |
246001_at |
AT5G20790
|
unknown |
0.314 |
0.056 |
| 68 |
261785_at |
AT1G08230
|
[Transmembrane amino acid transporter family protein] |
0.311 |
0.076 |
| 69 |
257635_at |
AT3G26280
|
CYP71B4, cytochrome P450, family 71, subfamily B, polypeptide 4 |
0.311 |
0.143 |
| 70 |
261023_at |
AT1G12200
|
FMO, flavin monooxygenase |
0.308 |
-0.078 |
| 71 |
264960_at |
AT1G76930
|
ATEXT1, EXTENSIN 1, ATEXT4, extensin 4, EXT1, extensin 1, EXT4, extensin 4, ORG5, OBP3-RESPONSIVE GENE 5 |
0.307 |
0.182 |
| 72 |
250499_at |
AT5G09730
|
ATBX3, ATBXL3, BETA-XYLOSIDASE 3, BX3, BXL3, beta-xylosidase 3, XYL3 |
0.305 |
0.119 |
| 73 |
253525_at |
AT4G31330
|
unknown |
0.302 |
-0.022 |
| 74 |
252859_at |
AT4G39780
|
[Integrase-type DNA-binding superfamily protein] |
0.302 |
0.096 |
| 75 |
261157_at |
AT1G34510
|
[Peroxidase superfamily protein] |
-1.090 |
-0.061 |
| 76 |
247871_at |
AT5G57530
|
AtXTH12, XTH12, xyloglucan endotransglucosylase/hydrolase 12 |
-1.009 |
-0.118 |
| 77 |
249934_at |
AT5G22410
|
RHS18, root hair specific 18 |
-0.949 |
-0.128 |
| 78 |
246652_at |
AT5G35190
|
EXT13, extensin 13 |
-0.905 |
-0.151 |
| 79 |
247914_at |
AT5G57540
|
AtXTH13, XTH13, xyloglucan endotransglucosylase/hydrolase 13 |
-0.866 |
-0.172 |
| 80 |
251226_at |
AT3G62680
|
ATPRP3, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 3, PRP3, proline-rich protein 3 |
-0.866 |
-0.087 |
| 81 |
252238_at |
AT3G49960
|
[Peroxidase superfamily protein] |
-0.823 |
-0.043 |
| 82 |
250801_at |
AT5G04960
|
[Plant invertase/pectin methylesterase inhibitor superfamily] |
-0.821 |
-0.211 |
| 83 |
254025_at |
AT4G25790
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.819 |
-0.135 |
| 84 |
253763_at |
AT4G28850
|
ATXTH26, XTH26, xyloglucan endotransglucosylase/hydrolase 26 |
-0.781 |
-0.156 |
| 85 |
263376_at |
AT2G20520
|
FLA6, FASCICLIN-like arabinogalactan 6 |
-0.775 |
-0.129 |
| 86 |
252833_at |
AT4G40090
|
AGP3, arabinogalactan protein 3 |
-0.774 |
-0.112 |
| 87 |
254044_at |
AT4G25820
|
ATXTH14, XTH14, xyloglucan endotransglucosylase/hydrolase 14, XTR9, xyloglucan endotransglycosylase 9 |
-0.773 |
-0.085 |
| 88 |
246991_at |
AT5G67400
|
RHS19, root hair specific 19 |
-0.748 |
-0.105 |
| 89 |
256352_at |
AT1G54970
|
ATPRP1, proline-rich protein 1, PRP1, proline-rich protein 1, RHS7, ROOT HAIR SPECIFIC 7 |
-0.689 |
-0.172 |
| 90 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.661 |
-0.057 |
| 91 |
255632_at |
AT4G00680
|
ADF8, actin depolymerizing factor 8 |
-0.659 |
-0.039 |
| 92 |
258765_at |
AT3G10710
|
RHS12, root hair specific 12 |
-0.643 |
-0.141 |
| 93 |
261562_at |
AT1G01750
|
ADF11, actin depolymerizing factor 11 |
-0.636 |
-0.130 |
| 94 |
254534_at |
AT4G19680
|
ATIRT2, IRON REGULATED TRANSPORTER 2, IRT2, iron regulated transporter 2 |
-0.617 |
-0.058 |
| 95 |
253613_at |
AT4G30320
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.614 |
-0.097 |
| 96 |
255516_at |
AT4G02270
|
RHS13, root hair specific 13 |
-0.589 |
-0.063 |
| 97 |
266276_at |
AT2G29330
|
TRI, tropinone reductase |
-0.586 |
-0.028 |
| 98 |
259525_at |
AT1G12560
|
ATEXP7, ATEXPA7, expansin A7, ATHEXP ALPHA 1.26, EXP7, EXPA7, expansin A7 |
-0.585 |
-0.051 |
| 99 |
261099_at |
AT1G62980
|
ATEXP18, ATEXPA18, expansin A18, ATHEXP ALPHA 1.25, EXP18, EXPANSIN 18, EXPA18, expansin A18 |
-0.559 |
-0.103 |
| 100 |
267191_at |
AT2G44110
|
ATMLO15, MILDEW RESISTANCE LOCUS O 15, MLO15, MILDEW RESISTANCE LOCUS O 15 |
-0.551 |
-0.123 |
| 101 |
246149_at |
AT5G19890
|
[Peroxidase superfamily protein] |
-0.544 |
0.084 |
| 102 |
260012_at |
AT1G67865
|
unknown |
-0.529 |
0.246 |
| 103 |
254107_at |
AT4G25220
|
AtG3Pp2, glycerol-3-phosphate permease 2, G3Pp2, glycerol-3-phosphate permease 2, RHS15, root hair specific 15 |
-0.509 |
-0.226 |
| 104 |
247094_at |
AT5G66280
|
GMD1, GDP-D-mannose 4,6-dehydratase 1 |
-0.506 |
-0.147 |
| 105 |
263614_at |
AT2G25240
|
AtCCP3, CCP3, conserved in ciliated species and in the land plants 3 |
-0.502 |
-0.062 |
| 106 |
265246_at |
AT2G43050
|
ATPMEPCRD |
-0.501 |
-0.262 |
| 107 |
245966_at |
AT5G19790
|
RAP2.11, related to AP2 11 |
-0.497 |
-0.134 |
| 108 |
266765_at |
AT2G46860
|
AtPPa3, pyrophosphorylase 3, PPa3, pyrophosphorylase 3 |
-0.478 |
-0.131 |
| 109 |
245506_at |
AT4G15700
|
[Thioredoxin superfamily protein] |
-0.475 |
0.068 |
| 110 |
251668_at |
AT3G57010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
-0.474 |
0.062 |
| 111 |
260758_at |
AT1G48930
|
AtGH9C1, glycosyl hydrolase 9C1, GH9C1, glycosyl hydrolase 9C1 |
-0.468 |
-0.056 |
| 112 |
250778_at |
AT5G05500
|
MOP10 |
-0.460 |
-0.084 |
| 113 |
245392_at |
AT4G15680
|
[Thioredoxin superfamily protein] |
-0.452 |
0.107 |
| 114 |
245736_at |
AT1G73330
|
ATDR4, drought-repressed 4, DR4, drought-repressed 4 |
-0.449 |
0.006 |
| 115 |
245967_at |
AT5G19800
|
[hydroxyproline-rich glycoprotein family protein] |
-0.445 |
-0.066 |
| 116 |
262603_at |
AT1G15380
|
GLYI4, glyoxylase I 4 |
-0.438 |
-0.090 |
| 117 |
258145_at |
AT3G18200
|
UMAMIT4, Usually multiple acids move in and out Transporters 4 |
-0.432 |
0.029 |
| 118 |
261991_at |
AT1G33700
|
[Beta-glucosidase, GBA2 type family protein] |
-0.432 |
-0.127 |
| 119 |
253301_at |
AT4G33720
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-0.431 |
0.097 |
| 120 |
260053_at |
AT1G78120
|
TPR12, tetratricopeptide repeat 12 |
-0.429 |
-0.021 |
| 121 |
249439_at |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
-0.427 |
0.009 |
| 122 |
253998_at |
AT4G26010
|
[Peroxidase superfamily protein] |
-0.425 |
-0.077 |
| 123 |
267094_at |
AT2G38080
|
ATLMCO4, ARABIDOPSIS LACCASE-LIKE MULTICOPPER OXIDASE 4, IRX12, IRREGULAR XYLEM 12, LAC4, LACCASE 4, LMCO4, LACCASE-LIKE MULTICOPPER OXIDASE 4 |
-0.424 |
-0.138 |
| 124 |
245504_at |
AT4G15660
|
[Thioredoxin superfamily protein] |
-0.416 |
0.111 |
| 125 |
265102_at |
AT1G30870
|
[Peroxidase superfamily protein] |
-0.413 |
-0.073 |
| 126 |
263552_x_at |
AT2G24980
|
EXT6, extensin 6 |
-0.410 |
-0.034 |
| 127 |
259548_at |
AT1G35260
|
MLP165, MLP-like protein 165 |
-0.409 |
-0.052 |
| 128 |
264647_at |
AT1G09090
|
ATRBOHB, respiratory burst oxidase homolog B, ATRBOHB-BETA, RBOHB, respiratory burst oxidase homolog B |
-0.407 |
-0.160 |
| 129 |
254891_at |
AT4G11740
|
SAY1 |
-0.401 |
-0.048 |
| 130 |
262883_at |
AT1G64780
|
AMT1;2, ammonium transporter 1;2, ATAMT1;2, ammonium transporter 1;2 |
-0.401 |
-0.199 |
| 131 |
257896_at |
AT3G16920
|
ATCTL2, CTL2, chitinase-like protein 2 |
-0.394 |
-0.062 |
| 132 |
264775_at |
AT1G22880
|
ATCEL5, ARABIDOPSIS THALIANA CELLULASE 5, ATGH9B4, ARABIDOPSIS THALIANA GLYCOSYL HYDROLASE 9B4, CEL5, cellulase 5 |
-0.391 |
0.076 |
| 133 |
267317_at |
AT2G34700
|
[Pollen Ole e 1 allergen and extensin family protein] |
-0.385 |
-0.120 |
| 134 |
253974_at |
AT4G26540
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.379 |
-0.087 |
| 135 |
253815_at |
AT4G28250
|
ATEXPB3, expansin B3, ATHEXP BETA 1.6, EXPB3, expansin B3 |
-0.377 |
-0.114 |
| 136 |
265031_at |
AT1G61590
|
[Protein kinase superfamily protein] |
-0.377 |
-0.141 |
| 137 |
248732_at |
AT5G48070
|
ATXTH20, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 20, XTH20, xyloglucan endotransglucosylase/hydrolase 20 |
-0.368 |
0.118 |
| 138 |
267457_at |
AT2G33790
|
AGP30, arabinogalactan protein 30, ATAGP30 |
-0.368 |
0.048 |
| 139 |
264347_at |
AT1G12040
|
LRX1, leucine-rich repeat/extensin 1 |
-0.366 |
-0.167 |
| 140 |
249996_at |
AT5G18600
|
[Thioredoxin superfamily protein] |
-0.362 |
0.079 |
| 141 |
245505_at |
AT4G15690
|
[Thioredoxin superfamily protein] |
-0.357 |
0.123 |
| 142 |
267595_at |
AT2G32990
|
AtGH9B8, glycosyl hydrolase 9B8, GH9B8, glycosyl hydrolase 9B8 |
-0.356 |
0.002 |
| 143 |
260344_at |
AT1G69240
|
ATMES15, ARABIDOPSIS THALIANA METHYL ESTERASE 15, MES15, methyl esterase 15, RHS9, ROOT HAIR SPECIFIC 9 |
-0.354 |
-0.176 |
| 144 |
251231_at |
AT3G62760
|
ATGSTF13 |
-0.353 |
-0.007 |
| 145 |
257761_at |
AT3G23090
|
WDL3 |
-0.350 |
-0.054 |
| 146 |
255876_at |
AT2G40480
|
unknown |
-0.330 |
0.014 |
| 147 |
261574_at |
AT1G01190
|
CYP78A8, cytochrome P450, family 78, subfamily A, polypeptide 8 |
-0.325 |
-0.054 |