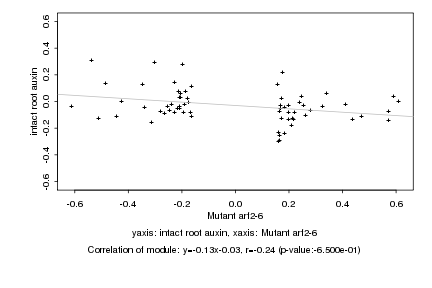
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant arf2-6 |
Log2 signal ratio
intact root auxin |
| 1 |
261046_at |
AT1G01390
|
[UDP-Glycosyltransferase superfamily protein] |
0.606 |
0.003 |
| 2 |
258498_at |
AT3G02480
|
[Late embryogenesis abundant protein (LEA) family protein] |
0.588 |
0.039 |
| 3 |
250868_at |
AT5G03860
|
MLS, malate synthase |
0.569 |
-0.138 |
| 4 |
249053_at |
AT5G44440
|
[FAD-binding Berberine family protein] |
0.569 |
-0.068 |
| 5 |
257947_at |
AT3G21720
|
ICL, isocitrate lyase |
0.469 |
-0.109 |
| 6 |
247486_at |
AT5G62140
|
unknown |
0.435 |
-0.132 |
| 7 |
253971_at |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
0.411 |
-0.017 |
| 8 |
256792_at |
AT3G22150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.337 |
0.064 |
| 9 |
262679_at |
AT1G75830
|
LCR67, low-molecular-weight cysteine-rich 67, PDF1.1, PLANT DEFENSIN 1.1 |
0.324 |
-0.035 |
| 10 |
245015_at |
ATCG00490
|
RBCL |
0.277 |
-0.062 |
| 11 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.260 |
-0.101 |
| 12 |
254663_at |
AT4G18290
|
KAT2, potassium channel in Arabidopsis thaliana 2 |
0.254 |
-0.023 |
| 13 |
246494_at |
AT5G16190
|
ATCSLA11, ARABIDOPSIS THALIANA CELLULOSE SYNTHASE LIKE A11, CSLA11, cellulose synthase like A11 |
0.245 |
0.045 |
| 14 |
247100_at |
AT5G66520
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.239 |
-0.007 |
| 15 |
264041_at |
AT2G03710
|
AGL3, AGAMOUS-like 3, SEP4, SEPALLATA 4 |
0.217 |
-0.077 |
| 16 |
251466_at |
AT3G59340
|
unknown |
0.216 |
-0.134 |
| 17 |
252317_at |
AT3G48720
|
DCF, DEFICIENT IN CUTIN FERULATE |
0.210 |
-0.127 |
| 18 |
247461_at |
AT5G62100
|
ATBAG2, BCL-2-associated athanogene 2, BAG2, BCL-2-associated athanogene 2 |
0.207 |
-0.173 |
| 19 |
267361_at |
AT2G39920
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.198 |
-0.082 |
| 20 |
247464_at |
AT5G62070
|
IQD23, IQ-domain 23 |
0.195 |
-0.129 |
| 21 |
244962_at |
ATCG01050
|
NDHD |
0.195 |
-0.027 |
| 22 |
245009_at |
ATCG00380
|
RPS4, chloroplast ribosomal protein S4 |
0.182 |
-0.043 |
| 23 |
262373_at |
AT1G73120
|
unknown |
0.180 |
-0.233 |
| 24 |
260237_at |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
0.174 |
0.224 |
| 25 |
249245_at |
AT5G42280
|
[Cysteine/Histidine-rich C1 domain family protein] |
0.171 |
-0.124 |
| 26 |
266279_at |
AT2G29290
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
0.170 |
0.029 |
| 27 |
265699_at |
AT2G03550
|
[alpha/beta-Hydrolases superfamily protein] |
0.166 |
-0.047 |
| 28 |
244998_at |
ATCG00180
|
RPOC1 |
0.165 |
-0.027 |
| 29 |
245006_at |
ATCG00340
|
PSAB |
0.164 |
-0.069 |
| 30 |
250549_at |
AT5G07860
|
[HXXXD-type acyl-transferase family protein] |
0.164 |
-0.292 |
| 31 |
267481_at |
AT2G02780
|
Leucine-rich repeat protein kinase family protein |
0.161 |
-0.249 |
| 32 |
267505_at |
AT2G45560
|
CYP76C1, cytochrome P450, family 76, subfamily C, polypeptide 1 |
0.160 |
-0.230 |
| 33 |
262830_at |
AT1G14700
|
ATPAP3, PAP3, purple acid phosphatase 3 |
0.160 |
-0.296 |
| 34 |
257744_at |
AT3G29230
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.154 |
0.134 |
| 35 |
249773_at |
AT5G24140
|
SQP2, squalene monooxygenase 2 |
-0.615 |
-0.037 |
| 36 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.540 |
0.310 |
| 37 |
250351_at |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-0.513 |
-0.123 |
| 38 |
261648_at |
AT1G27730
|
STZ, salt tolerance zinc finger, ZAT10 |
-0.487 |
0.136 |
| 39 |
246001_at |
AT5G20790
|
unknown |
-0.446 |
-0.112 |
| 40 |
263391_at |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-0.429 |
0.005 |
| 41 |
249675_at |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-0.349 |
0.134 |
| 42 |
264872_at |
AT1G24260
|
AGL9, AGAMOUS-like 9, SEP3, SEPALLATA3 |
-0.341 |
-0.039 |
| 43 |
248729_at |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
-0.316 |
-0.154 |
| 44 |
263402_at |
AT2G04050
|
[MATE efflux family protein] |
-0.303 |
0.298 |
| 45 |
252234_at |
AT3G49780
|
ATPSK3 (FORMER SYMBOL), ATPSK4, phytosulfokine 4 precursor, PSK4, phytosulfokine 4 precursor |
-0.282 |
-0.069 |
| 46 |
254059_at |
AT4G25200
|
ATHSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6, HSP23.6-MITO, mitochondrion-localized small heat shock protein 23.6 |
-0.265 |
-0.083 |
| 47 |
249545_at |
AT5G38030
|
[MATE efflux family protein] |
-0.256 |
-0.033 |
| 48 |
258930_at |
AT3G10040
|
[sequence-specific DNA binding transcription factors] |
-0.247 |
-0.063 |
| 49 |
259846_at |
AT1G72140
|
[Major facilitator superfamily protein] |
-0.242 |
-0.017 |
| 50 |
256442_at |
AT3G10930
|
unknown |
-0.230 |
0.145 |
| 51 |
263208_at |
AT1G10480
|
ZFP5, zinc finger protein 5 |
-0.229 |
-0.076 |
| 52 |
256940_at |
AT3G30720
|
QQS, QUA-QUINE STARCH |
-0.217 |
-0.046 |
| 53 |
247691_at |
AT5G59720
|
HSP18.2, heat shock protein 18.2 |
-0.214 |
0.078 |
| 54 |
263594_at |
AT2G01880
|
ATPAP7, PURPLE ACID PHOSPHATASE 7, PAP7, purple acid phosphatase 7 |
-0.212 |
-0.050 |
| 55 |
266132_at |
AT2G45130
|
ATSPX3, ARABIDOPSIS THALIANA SPX DOMAIN GENE 3, SPX3, SPX domain gene 3 |
-0.209 |
-0.030 |
| 56 |
262616_at |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.209 |
0.032 |
| 57 |
262887_at |
AT1G14780
|
[MAC/Perforin domain-containing protein] |
-0.208 |
0.062 |
| 58 |
266992_at |
AT2G39200
|
ATMLO12, MILDEW RESISTANCE LOCUS O 12, MLO12, MILDEW RESISTANCE LOCUS O 12 |
-0.206 |
0.033 |
| 59 |
258887_at |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-0.201 |
0.283 |
| 60 |
264859_at |
AT1G24280
|
G6PD3, glucose-6-phosphate dehydrogenase 3 |
-0.196 |
-0.080 |
| 61 |
246093_at |
AT5G20550
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.192 |
-0.016 |
| 62 |
257774_at |
AT3G29250
|
AtSDR4, SDR4, short-chain dehydrogenase reductase 4 |
-0.189 |
0.077 |
| 63 |
252539_at |
AT3G45730
|
unknown |
-0.182 |
0.030 |
| 64 |
254574_at |
AT4G19430
|
unknown |
-0.179 |
-0.006 |
| 65 |
259015_at |
AT3G07350
|
unknown |
-0.169 |
-0.078 |
| 66 |
254839_at |
AT4G12400
|
Hop3, Hop3 |
-0.166 |
0.113 |
| 67 |
266259_at |
AT2G27830
|
unknown |
-0.165 |
-0.105 |