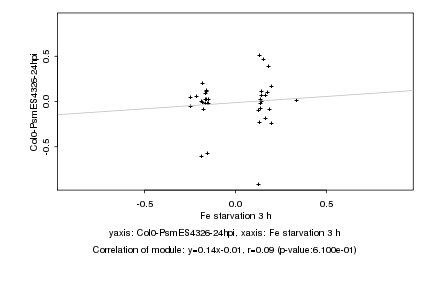
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Fe starvation 3 h |
Log2 signal ratio
Col0-PsmES4326-24hpi |
| 1 |
247070_at |
AT5G66815
|
unknown |
0.335 |
0.021 |
| 2 |
263020_at |
AT1G23880
|
[NHL domain-containing protein] |
0.198 |
-0.240 |
| 3 |
263198_at |
AT1G53990
|
GLIP3, GDSL-motif lipase 3 |
0.194 |
0.168 |
| 4 |
246733_at |
AT5G27660
|
DEG14, degradation of periplasmic proteins 14 |
0.182 |
-0.079 |
| 5 |
255568_at |
AT4G01250
|
AtWRKY22, WRKY22 |
0.177 |
0.389 |
| 6 |
262018_at |
AT1G35617
|
unknown |
0.176 |
0.107 |
| 7 |
255870_at |
AT2G30280
|
DMS4, DEFECTIVE IN MERISTEM SILENCING 4, RDM4, RNA-directed DNA methylation 4 |
0.165 |
-0.180 |
| 8 |
267445_at |
AT2G33680
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
0.161 |
0.076 |
| 9 |
257401_at |
AT1G23550
|
SRO2, similar to RCD one 2 |
0.152 |
0.466 |
| 10 |
255022_at |
AT4G10350
|
ANAC070, NAC domain containing protein 70, BRN2, BEARSKIN 2, NAC070, NAC domain containing protein 70 |
0.141 |
0.002 |
| 11 |
253045_at |
AT4G37445
|
unknown |
0.141 |
0.067 |
| 12 |
250374_at |
AT5G11530
|
EMF1, embryonic flower 1 |
0.138 |
0.116 |
| 13 |
248570_at |
AT5G49780
|
[Leucine-rich repeat protein kinase family protein] |
0.137 |
0.031 |
| 14 |
252650_at |
AT3G44690
|
unknown |
0.133 |
-0.016 |
| 15 |
254877_at |
AT4G11640
|
ATSR, serine racemase, SR, serine racemase |
0.133 |
-0.074 |
| 16 |
267028_at |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
0.132 |
0.515 |
| 17 |
263474_at |
AT2G31725
|
unknown |
0.130 |
-0.231 |
| 18 |
248605_at |
AT5G49410
|
unknown |
0.125 |
-0.093 |
| 19 |
262682_at |
AT1G75900
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.124 |
-0.906 |
| 20 |
256003_at |
AT3G31430
|
unknown |
-0.251 |
0.047 |
| 21 |
254064_at |
AT4G25410
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.250 |
-0.050 |
| 22 |
264129_at |
AT1G79170
|
unknown |
-0.218 |
0.060 |
| 23 |
246727_at |
AT5G28010
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.190 |
0.006 |
| 24 |
250884_at |
AT5G03940
|
54CP, 54 CHLOROPLAST PROTEIN, CPSRP54, chloroplast signal recognition particle 54 kDa subunit, FFC, FIFTY-FOUR CHLOROPLAST HOMOLOGUE, SRP54CP, SIGNAL RECOGNITION PARTICLE 54 KDA SUBUNIT CHLOROPLAST PROTEIN |
-0.189 |
-0.602 |
| 25 |
254886_at |
AT4G11760
|
LCR17, low-molecular-weight cysteine-rich 17 |
-0.186 |
-0.002 |
| 26 |
258762_at |
AT3G10750
|
[FBD domain family] |
-0.183 |
0.202 |
| 27 |
260070_at |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.176 |
-0.087 |
| 28 |
262359_at |
AT1G73066
|
[Leucine-rich repeat family protein] |
-0.169 |
0.031 |
| 29 |
266594_at |
AT2G46190
|
[Mitochondrial glycoprotein family protein] |
-0.167 |
0.093 |
| 30 |
245019_at |
ATCG00530
|
YCF10 |
-0.165 |
-0.015 |
| 31 |
260103_at |
AT1G35430
|
unknown |
-0.163 |
0.030 |
| 32 |
246617_at |
AT5G36270
|
[similar to DHAR2, glutathione dehydrogenase (ascorbate) [Arabidopsis thaliana] (TAIR:AT1G75270.1)] |
-0.163 |
0.131 |
| 33 |
248928_at |
AT5G45970
|
ARAC2, Arabidopsis RAC-like 2, ATRAC2, ARABIDOPSIS THALIANA RAC 2, ATROP7, RHO-RELATED PROTEIN FROM PLANTS 7, RAC2, RAC-like 2, ROP7 |
-0.161 |
0.120 |
| 34 |
250696_at |
AT5G06790
|
unknown |
-0.158 |
-0.567 |
| 35 |
253734_at |
AT4G29180
|
RHS16, root hair specific 16 |
-0.155 |
-0.020 |
| 36 |
252333_at |
AT3G48830
|
[polynucleotide adenylyltransferase family protein / RNA recognition motif (RRM)-containing protein] |
-0.153 |
-0.020 |
| 37 |
258066_at |
AT3G25970
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-0.151 |
0.026 |