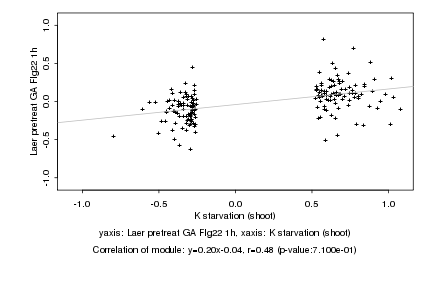
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
K starvation (shoot) |
Log2 signal ratio
Laer pretreat GA Flg22 1h |
| 1 |
252136_at |
AT3G50770
|
CML41, calmodulin-like 41 |
1.076 |
-0.099 |
| 2 |
258947_at |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
1.030 |
0.055 |
| 3 |
248333_at |
AT5G52390
|
[PAR1 protein] |
1.017 |
0.312 |
| 4 |
254101_at |
AT4G25000
|
AMY1, alpha-amylase-like, ATAMY1 |
1.007 |
-0.299 |
| 5 |
260881_at |
AT1G21550
|
[Calcium-binding EF-hand family protein] |
0.979 |
0.094 |
| 6 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
0.947 |
0.008 |
| 7 |
252102_at |
AT3G50970
|
LTI30, LOW TEMPERATURE-INDUCED 30, XERO2 |
0.925 |
-0.086 |
| 8 |
245840_at |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
0.906 |
0.298 |
| 9 |
264580_at |
AT1G05340
|
unknown |
0.892 |
0.138 |
| 10 |
250090_at |
AT5G17330
|
GAD, glutamate decarboxylase, GAD1, GLUTAMATE DECARBOXYLASE 1 |
0.878 |
0.521 |
| 11 |
249626_at |
AT5G37540
|
[Eukaryotic aspartyl protease family protein] |
0.873 |
-0.054 |
| 12 |
266545_at |
AT2G35290
|
SAUR79, SMALL AUXIN UPREGULATED RNA 79 |
0.843 |
0.234 |
| 13 |
265853_at |
AT2G42360
|
[RING/U-box superfamily protein] |
0.838 |
0.205 |
| 14 |
260804_at |
AT1G78410
|
[VQ motif-containing protein] |
0.831 |
-0.314 |
| 15 |
249940_at |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
0.820 |
0.093 |
| 16 |
259987_at |
AT1G75030
|
ATLP-3, thaumatin-like protein 3, TLP-3, thaumatin-like protein 3 |
0.801 |
0.051 |
| 17 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
0.799 |
0.077 |
| 18 |
254667_at |
AT4G18280
|
[glycine-rich cell wall protein-related] |
0.790 |
-0.301 |
| 19 |
247095_at |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
0.784 |
0.118 |
| 20 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.783 |
0.222 |
| 21 |
259850_at |
AT1G72240
|
unknown |
0.777 |
0.064 |
| 22 |
256012_at |
AT1G19250
|
FMO1, flavin-dependent monooxygenase 1 |
0.768 |
0.700 |
| 23 |
261366_at |
AT1G53100
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
0.762 |
0.111 |
| 24 |
254490_at |
AT4G20320
|
[CTP synthase family protein] |
0.761 |
0.147 |
| 25 |
246842_at |
AT5G26731
|
unknown |
0.745 |
0.112 |
| 26 |
250344_at |
AT5G11930
|
[Thioredoxin superfamily protein] |
0.743 |
0.019 |
| 27 |
258203_at |
AT3G13950
|
unknown |
0.739 |
0.191 |
| 28 |
248466_at |
AT5G50720
|
ATHVA22E, ARABIDOPSIS THALIANA HVA22 HOMOLOGUE E, HVA22E, HVA22 homologue E |
0.735 |
-0.051 |
| 29 |
262930_at |
AT1G65690
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.734 |
0.376 |
| 30 |
255941_at |
AT1G20350
|
ATTIM17-1, translocase inner membrane subunit 17-1, TIM17-1, translocase inner membrane subunit 17-1 |
0.715 |
0.169 |
| 31 |
262518_at |
AT1G17170
|
ATGSTU24, glutathione S-transferase TAU 24, GST, Arabidopsis thaliana Glutathione S-transferase (class tau) 24, GSTU24, glutathione S-transferase TAU 24 |
0.708 |
0.073 |
| 32 |
246272_at |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
0.699 |
0.027 |
| 33 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
0.696 |
0.275 |
| 34 |
260005_at |
AT1G67920
|
unknown |
0.689 |
0.164 |
| 35 |
260386_at |
AT1G74010
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.686 |
0.102 |
| 36 |
252437_at |
AT3G47380
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.679 |
0.097 |
| 37 |
265214_at |
AT1G05000
|
AtPFA-DSP1, PFA-DSP1, plant and fungi atypical dual-specificity phosphatase 1 |
0.678 |
0.248 |
| 38 |
253632_at |
AT4G30430
|
TET9, tetraspanin9 |
0.677 |
0.274 |
| 39 |
247487_at |
AT5G62150
|
[peptidoglycan-binding LysM domain-containing protein] |
0.673 |
0.300 |
| 40 |
252068_at |
AT3G51440
|
[Calcium-dependent phosphotriesterase superfamily protein] |
0.673 |
0.091 |
| 41 |
258209_at |
AT3G14060
|
unknown |
0.672 |
-0.088 |
| 42 |
250286_at |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
0.664 |
0.349 |
| 43 |
266778_at |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
0.663 |
-0.439 |
| 44 |
260405_at |
AT1G69930
|
ATGSTU11, glutathione S-transferase TAU 11, GSTU11, glutathione S-transferase TAU 11 |
0.658 |
0.089 |
| 45 |
256252_at |
AT3G11340
|
UGT76B1, UDP-dependent glycosyltransferase 76B1 |
0.657 |
0.121 |
| 46 |
248819_at |
AT5G47050
|
[SBP (S-ribonuclease binding protein) family protein] |
0.657 |
0.090 |
| 47 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
0.652 |
-0.018 |
| 48 |
246125_at |
AT5G19875
|
unknown |
0.652 |
-0.219 |
| 49 |
250983_at |
AT5G02780
|
GSTL1, glutathione transferase lambda 1 |
0.648 |
0.445 |
| 50 |
264866_at |
AT1G24140
|
[Matrixin family protein] |
0.646 |
0.096 |
| 51 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
0.644 |
0.212 |
| 52 |
255630_at |
AT4G00700
|
[C2 calcium/lipid-binding plant phosphoribosyltransferase family protein] |
0.641 |
0.029 |
| 53 |
264400_at |
AT1G61800
|
ATGPT2, ARABIDOPSIS GLUCOSE-6-PHOSPHATE/PHOSPHATE TRANSLOCATOR 2, GPT2, glucose-6-phosphate/phosphate translocator 2 |
0.638 |
0.268 |
| 54 |
253608_at |
AT4G30290
|
ATXTH19, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 19, XTH19, xyloglucan endotransglucosylase/hydrolase 19 |
0.635 |
0.115 |
| 55 |
267565_at |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
0.634 |
0.510 |
| 56 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
0.627 |
0.280 |
| 57 |
264758_at |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
0.627 |
0.200 |
| 58 |
264729_at |
AT1G22990
|
HIPP22, heavy metal associated isoprenylated plant protein 22 |
0.624 |
0.095 |
| 59 |
246302_at |
AT3G51860
|
ATCAX3, ATHCX1, CAX1-LIKE, CAX3, cation exchanger 3 |
0.623 |
-0.172 |
| 60 |
263584_at |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
0.620 |
0.074 |
| 61 |
256877_at |
AT3G26470
|
[Powdery mildew resistance protein, RPW8 domain] |
0.617 |
0.020 |
| 62 |
252214_at |
AT3G50260
|
ATERF#011, CEJ1, cooperatively regulated by ethylene and jasmonate 1, DEAR1, DREB AND EAR MOTIF PROTEIN 1 |
0.613 |
0.192 |
| 63 |
259410_at |
AT1G13340
|
[Regulator of Vps4 activity in the MVB pathway protein] |
0.609 |
0.299 |
| 64 |
261033_at |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
0.607 |
0.025 |
| 65 |
254178_at |
AT4G23880
|
unknown |
0.595 |
-0.110 |
| 66 |
265725_at |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
0.594 |
0.034 |
| 67 |
266799_at |
AT2G22860
|
ATPSK2, phytosulfokine 2 precursor, PSK2, phytosulfokine 2 precursor |
0.594 |
0.085 |
| 68 |
265672_at |
AT2G31980
|
AtCYS2, PHYTOCYSTATIN 2, CYS2, PHYTOCYSTATIN 2 |
0.592 |
0.087 |
| 69 |
262229_at |
AT1G68620
|
[alpha/beta-Hydrolases superfamily protein] |
0.591 |
0.137 |
| 70 |
247109_at |
AT5G65870
|
ATPSK5, phytosulfokine 5 precursor, PSK5, PSK5, phytosulfokine 5 precursor |
0.583 |
-0.507 |
| 71 |
258322_at |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
0.580 |
-0.103 |
| 72 |
251176_at |
AT3G63380
|
[ATPase E1-E2 type family protein / haloacid dehalogenase-like hydrolase family protein] |
0.578 |
0.136 |
| 73 |
257644_at |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
0.577 |
0.116 |
| 74 |
262482_at |
AT1G17020
|
ATSRG1, SENESCENCE-RELATED GENE 1, SRG1, senescence-related gene 1 |
0.577 |
-0.061 |
| 75 |
256647_at |
AT3G13610
|
2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein |
0.569 |
0.816 |
| 76 |
259655_at |
AT1G55210
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
0.566 |
0.070 |
| 77 |
250724_at |
AT5G06330
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
0.561 |
0.089 |
| 78 |
256114_at |
AT1G16850
|
unknown |
0.560 |
0.156 |
| 79 |
250646_at |
AT5G06720
|
ATPA2, peroxidase 2, AtPRX53, PA2, peroxidase 2, PRX53, peroxidase 53 |
0.560 |
0.244 |
| 80 |
249096_at |
AT5G43910
|
[pfkB-like carbohydrate kinase family protein] |
0.560 |
0.211 |
| 81 |
262703_at |
AT1G16510
|
SAUR41, SMALL AUXIN UPREGULATED 41 |
0.552 |
-0.197 |
| 82 |
251137_at |
AT5G01300
|
[PEBP (phosphatidylethanolamine-binding protein) family protein] |
0.550 |
0.005 |
| 83 |
246821_at |
AT5G26920
|
CBP60G, Cam-binding protein 60-like G |
0.549 |
0.389 |
| 84 |
263411_at |
AT2G28710
|
[C2H2-type zinc finger family protein] |
0.545 |
0.056 |
| 85 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
0.543 |
0.128 |
| 86 |
256839_at |
AT3G22930
|
AtCML11, CML11, calmodulin-like 11 |
0.539 |
-0.222 |
| 87 |
260581_at |
AT2G47190
|
ATMYB2, MYB DOMAIN PROTEIN 2, MYB2, myb domain protein 2 |
0.538 |
0.091 |
| 88 |
266462_at |
AT2G47770
|
ATTSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related, TSPO, TSPO(outer membrane tryptophan-rich sensory protein)-related |
0.535 |
-0.074 |
| 89 |
247047_at |
AT5G66650
|
unknown |
0.534 |
0.168 |
| 90 |
245982_at |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
0.527 |
0.153 |
| 91 |
247723_at |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
0.525 |
0.162 |
| 92 |
252421_at |
AT3G47540
|
[Chitinase family protein] |
0.525 |
0.204 |
| 93 |
259507_at |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
0.520 |
0.041 |
| 94 |
257964_at |
AT3G19850
|
[Phototropic-responsive NPH3 family protein] |
-0.801 |
-0.457 |
| 95 |
261873_at |
AT1G11350
|
CBRLK1, CALMODULIN-BINDING RECEPTOR-LIKE PROTEIN KINASE, RKS2, SD1-13, S-domain-1 13 |
-0.611 |
-0.094 |
| 96 |
253423_at |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.562 |
-0.008 |
| 97 |
248895_at |
AT5G46330
|
FLS2, FLAGELLIN-SENSITIVE 2 |
-0.523 |
-0.012 |
| 98 |
257895_at |
AT3G16950
|
LPD1, lipoamide dehydrogenase 1, ptlpd1 |
-0.507 |
-0.412 |
| 99 |
251013_at |
AT5G02540
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
-0.486 |
-0.254 |
| 100 |
247780_at |
AT5G58770
|
AtcPT4, cPT4, cis-prenyltransferase 4 |
-0.462 |
-0.259 |
| 101 |
251017_at |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-0.452 |
-0.119 |
| 102 |
251402_at |
AT3G60290
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.452 |
-0.131 |
| 103 |
247348_at |
AT5G63810
|
AtBGAL10, Arabidopsis thaliana beta-galactosidase 10, BGAL10, beta-galactosidase 10 |
-0.444 |
0.008 |
| 104 |
255652_at |
AT4G00950
|
MEE47, maternal effect embryo arrest 47 |
-0.437 |
-0.089 |
| 105 |
265724_at |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-0.432 |
0.024 |
| 106 |
256060_at |
AT1G07050
|
[CCT motif family protein] |
-0.420 |
0.168 |
| 107 |
261043_at |
AT1G01220
|
AtFKGP, Arabidopsis thaliana L-fucokinase/GDP-L-fucose pyrophosphorylase, FKGP, L-fucokinase/GDP-L-fucose pyrophosphorylase |
-0.416 |
0.106 |
| 108 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
-0.413 |
-0.051 |
| 109 |
258708_at |
AT3G09580
|
[FAD/NAD(P)-binding oxidoreductase family protein] |
-0.412 |
-0.371 |
| 110 |
267556_at |
AT2G32810
|
BGAL9, beta galactosidase 9, BGAL9, beta-galactosidase 9 |
-0.410 |
-0.130 |
| 111 |
248624_at |
AT5G48790
|
unknown |
-0.403 |
-0.127 |
| 112 |
254553_at |
AT4G19530
|
[disease resistance protein (TIR-NBS-LRR class) family] |
-0.403 |
-0.488 |
| 113 |
246483_at |
AT5G16000
|
AtNIK1, NIK1, NSP-interacting kinase 1 |
-0.403 |
0.016 |
| 114 |
265414_at |
AT2G16660
|
[Major facilitator superfamily protein] |
-0.395 |
-0.139 |
| 115 |
254218_at |
AT4G23740
|
[Leucine-rich repeat protein kinase family protein] |
-0.392 |
-0.276 |
| 116 |
248404_at |
AT5G51460
|
ATTPPA, TPPA, trehalose-6-phosphate phosphatase A |
-0.382 |
-0.152 |
| 117 |
264508_at |
AT1G09570
|
FHY2, FAR RED ELONGATED HYPOCOTYL 2, FRE1, FAR RED ELONGATED 1, HY8, ELONGATED HYPOCOTYL 8, PHYA, phytochrome A |
-0.379 |
-0.151 |
| 118 |
248420_at |
AT5G51560
|
[Leucine-rich repeat protein kinase family protein] |
-0.378 |
-0.065 |
| 119 |
251629_at |
AT3G57410
|
ATVLN3, VLN3, villin 3 |
-0.376 |
0.003 |
| 120 |
263883_at |
AT2G21830
|
[Cysteine/Histidine-rich C1 domain family protein] |
-0.373 |
-0.037 |
| 121 |
250613_at |
AT5G07240
|
IQD24, IQ-domain 24 |
-0.372 |
-0.568 |
| 122 |
246371_at |
AT1G51940
|
LYK3, LysM-containing receptor-like kinase 3 |
-0.367 |
-0.185 |
| 123 |
248994_at |
AT5G45250
|
RPS4, RESISTANT TO P. SYRINGAE 4 |
-0.362 |
-0.016 |
| 124 |
260974_at |
AT1G53440
|
[Leucine-rich repeat transmembrane protein kinase] |
-0.360 |
0.124 |
| 125 |
263499_at |
AT2G42580
|
TTL3, tetratricopetide-repeat thioredoxin-like 3, VIT, VHI-INTERACTING TPR CONTAINING PROTEIN |
-0.359 |
-0.045 |
| 126 |
256772_at |
AT3G13750
|
BGAL1, beta galactosidase 1, BGAL1, beta-galactosidase 1 |
-0.348 |
-0.350 |
| 127 |
261395_at |
AT1G79700
|
WRI4, WRINKLED 4 |
-0.345 |
-0.095 |
| 128 |
259104_at |
AT3G02170
|
LNG2, LONGIFOLIA2, TRM1, TON1 Recruiting Motif 1 |
-0.343 |
-0.041 |
| 129 |
253947_at |
AT4G26760
|
MAP65-2, microtubule-associated protein 65-2 |
-0.343 |
0.058 |
| 130 |
261926_at |
AT1G22530
|
PATL2, PATELLIN 2 |
-0.343 |
-0.190 |
| 131 |
250885_at |
AT5G03910
|
ABCB29, ATP-binding cassette B29, ATATH12, ARABIDOPSIS THALIANA ABC TRANSPORTER HOMOLOG 12, ATH12, ABC2 homolog 12 |
-0.341 |
-0.014 |
| 132 |
263612_at |
AT2G16440
|
MCM4, MINICHROMOSOME MAINTENANCE 4 |
-0.330 |
0.245 |
| 133 |
264990_at |
AT1G27210
|
[ARM repeat superfamily protein] |
-0.327 |
0.119 |
| 134 |
256503_at |
AT1G75250
|
ATRL6, RAD-like 6, RL6, RAD-like 6, RSM3, RADIALIS-LIKE SANT/MYB 3 |
-0.324 |
-0.194 |
| 135 |
258156_at |
AT3G18050
|
unknown |
-0.323 |
-0.279 |
| 136 |
267606_at |
AT2G26640
|
KCS11, 3-ketoacyl-CoA synthase 11 |
-0.323 |
-0.380 |
| 137 |
264568_at |
AT1G05150
|
[Calcium-binding tetratricopeptide family protein] |
-0.322 |
0.093 |
| 138 |
258006_at |
AT3G19400
|
[Cysteine proteinases superfamily protein] |
-0.320 |
0.071 |
| 139 |
267465_at |
AT2G19170
|
SLP3, subtilisin-like serine protease 3 |
-0.319 |
0.028 |
| 140 |
251996_at |
AT3G52840
|
BGAL2, beta-galactosidase 2 |
-0.319 |
-0.239 |
| 141 |
264521_at |
AT1G10020
|
unknown |
-0.318 |
-0.041 |
| 142 |
260041_at |
AT1G68780
|
[RNI-like superfamily protein] |
-0.313 |
0.077 |
| 143 |
257651_at |
AT3G16850
|
[Pectin lyase-like superfamily protein] |
-0.309 |
-0.169 |
| 144 |
245696_at |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-0.307 |
-0.234 |
| 145 |
254221_at |
AT4G23820
|
[Pectin lyase-like superfamily protein] |
-0.306 |
-0.303 |
| 146 |
255540_at |
AT4G01800
|
AGY1, Albino or Glassy Yellow 1, AtcpSecA, Arabidopsis thaliana chloroplast SecA, SECA1 |
-0.299 |
-0.244 |
| 147 |
253337_at |
AT4G33470
|
ATHDA14, HDA14, histone deacetylase 14 |
-0.299 |
-0.291 |
| 148 |
260603_at |
AT1G55960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.298 |
-0.241 |
| 149 |
257772_at |
AT3G23080
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-0.298 |
-0.060 |
| 150 |
267310_at |
AT2G34680
|
AIR9, AUXIN-INDUCED IN ROOT CULTURES 9 |
-0.297 |
-0.172 |
| 151 |
266302_at |
AT2G27050
|
AtEIL1, EIL1, ETHYLENE-INSENSITIVE3-like 1 |
-0.296 |
-0.146 |
| 152 |
252543_at |
AT3G45780
|
JK224, NPH1, NONPHOTOTROPIC HYPOCOTYL 1, PHOT1, phototropin 1, RPT1, ROOT PHOTOTROPISM 1 |
-0.295 |
-0.621 |
| 153 |
248177_at |
AT5G54630
|
[zinc finger protein-related] |
-0.294 |
-0.188 |
| 154 |
260152_at |
AT1G52830
|
IAA6, indole-3-acetic acid 6, SHY1, SHORT HYPOCOTYL 1 |
-0.293 |
0.075 |
| 155 |
263957_at |
AT2G35880
|
[TPX2 (targeting protein for Xklp2) protein family] |
-0.292 |
-0.156 |
| 156 |
261783_at |
AT1G08190
|
ATVAM2, ATVPS41, VACUOLAR PROTEIN SORTING 41, VAM2, VPS41, vacuolar protein sorting 41, ZIP2, ZIGZAG SUPPRESSOR 2 |
-0.292 |
-0.007 |
| 157 |
249354_at |
AT5G40480
|
EMB3012, embryo defective 3012 |
-0.292 |
-0.153 |
| 158 |
260465_at |
AT1G10910
|
EMB3103, EMBRYO DEFECTIVE 3103 |
-0.292 |
-0.235 |
| 159 |
261801_at |
AT1G30520
|
AAE14, acyl-activating enzyme 14 |
-0.292 |
-0.262 |
| 160 |
258537_at |
AT3G04210
|
[Disease resistance protein (TIR-NBS class)] |
-0.289 |
-0.225 |
| 161 |
253533_at |
AT4G31590
|
ATCSLC05, CELLULOSE-SYNTHASE LIKE C5, ATCSLC5, Cellulose-synthase-like C5, CSLC05, CELLULOSE-SYNTHASE LIKE C5, CSLC5, Cellulose-synthase-like C5 |
-0.287 |
-0.042 |
| 162 |
245176_at |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.286 |
-0.077 |
| 163 |
259083_at |
AT3G04810
|
ATNEK2, NIMA-related kinase 2, NEK2, NIMA-related kinase 2 |
-0.286 |
-0.108 |
| 164 |
254815_at |
AT4G12420
|
SKU5 |
-0.284 |
-0.104 |
| 165 |
249742_at |
AT5G24490
|
[30S ribosomal protein, putative] |
-0.283 |
-0.143 |
| 166 |
261834_at |
AT1G10640
|
[Pectin lyase-like superfamily protein] |
-0.283 |
0.045 |
| 167 |
266642_at |
AT2G35410
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.282 |
-0.107 |
| 168 |
266489_at |
AT2G35190
|
ATNPSN11, NPSN11, novel plant snare 11, NSPN11 |
-0.281 |
0.452 |
| 169 |
263558_at |
AT2G16380
|
[Sec14p-like phosphatidylinositol transfer family protein] |
-0.280 |
-0.065 |
| 170 |
250102_at |
AT5G16590
|
LRR1, Leucine rich repeat protein 1 |
-0.280 |
-0.277 |
| 171 |
263805_at |
AT2G40400
|
unknown |
-0.280 |
-0.013 |
| 172 |
254089_at |
AT4G24800
|
ECIP1, EIN2 C-terminus Interacting Protein 1 |
-0.276 |
0.021 |
| 173 |
265066_at |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-0.276 |
-0.049 |
| 174 |
255513_at |
AT4G02060
|
MCM7, PRL, PROLIFERA |
-0.274 |
0.217 |
| 175 |
267532_at |
AT2G42040
|
unknown |
-0.274 |
0.098 |
| 176 |
266509_at |
AT2G47940
|
DEG2, degradation of periplasmic proteins 2, DEGP2, DEGP protease 2, EMB3117, EMBRYO DEFECTIVE 3117 |
-0.273 |
-0.317 |
| 177 |
261451_at |
AT1G21060
|
unknown |
-0.272 |
-0.161 |
| 178 |
246288_at |
AT1G31850
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.272 |
0.156 |
| 179 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.270 |
-0.108 |
| 180 |
266899_at |
AT2G34620
|
[Mitochondrial transcription termination factor family protein] |
-0.266 |
-0.397 |
| 181 |
263163_at |
AT1G03160
|
FZL, FZO-like |
-0.264 |
-0.096 |
| 182 |
249327_at |
AT5G40890
|
ATCLC-A, chloride channel A, ATCLCA, CLC-A, chloride channel A, CLC-A, CHLORIDE CHANNEL-A, CLCA, CHLORIDE CHANNEL A |
-0.262 |
-0.206 |
| 183 |
255617_at |
AT4G01330
|
[Protein kinase superfamily protein] |
-0.261 |
-0.298 |
| 184 |
251261_at |
AT3G62110
|
[Pectin lyase-like superfamily protein] |
-0.259 |
0.038 |
| 185 |
264783_at |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.257 |
-0.030 |