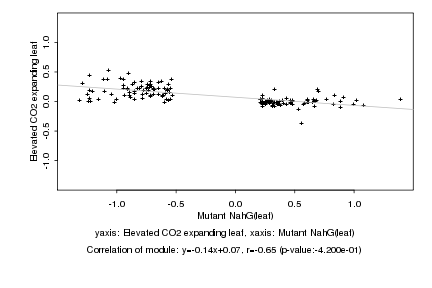
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
Mutant NahG(leaf) |
Log2 signal ratio
Elevated CO2 expanding leaf |
| 1 |
260221_at |
AT1G74670
|
GASA6, GA-stimulated Arabidopsis 6 |
1.388 |
0.042 |
| 2 |
265588_at |
AT2G19970
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
1.075 |
-0.065 |
| 3 |
263161_at |
AT1G54020
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.020 |
0.023 |
| 4 |
265586_at |
AT2G19990
|
PR-1-LIKE, pathogenesis-related protein-1-like |
0.994 |
-0.035 |
| 5 |
255524_at |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.906 |
0.085 |
| 6 |
256527_at |
AT1G66100
|
[Plant thionin] |
0.885 |
0.015 |
| 7 |
245422_at |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
0.879 |
-0.093 |
| 8 |
259802_at |
AT1G72260
|
THI2.1, thionin 2.1, THI2.1.1 |
0.831 |
0.109 |
| 9 |
252639_at |
AT3G44550
|
FAR5, fatty acid reductase 5 |
0.819 |
-0.047 |
| 10 |
256577_at |
AT3G28220
|
[TRAF-like family protein] |
0.764 |
0.039 |
| 11 |
257365_x_at |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
0.693 |
0.173 |
| 12 |
249052_at |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
0.687 |
0.206 |
| 13 |
245275_at |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
0.679 |
0.020 |
| 14 |
266215_at |
AT2G06850
|
EXGT-A1, endoxyloglucan transferase A1, EXT, ENDOXYLOGLUCAN TRANSFERASE, XTH4, xyloglucan endotransglucosylase/hydrolase 4 |
0.671 |
0.012 |
| 15 |
256299_at |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
0.666 |
0.005 |
| 16 |
259009_at |
AT3G09260
|
BGLU23, LEB, LONG ER BODY, PSR3.1, PYK10 |
0.662 |
-0.070 |
| 17 |
265083_at |
AT1G03820
|
unknown |
0.655 |
0.044 |
| 18 |
254726_at |
AT4G13660
|
ATPRR2, PRR2, pinoresinol reductase 2 |
0.647 |
0.000 |
| 19 |
263431_at |
AT2G22170
|
PLAT2, PLAT domain protein 2 |
0.613 |
-0.011 |
| 20 |
247573_at |
AT5G61160
|
AACT1, anthocyanin 5-aromatic acyltransferase 1 |
0.606 |
0.045 |
| 21 |
262643_at |
AT1G62770
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
0.606 |
-0.008 |
| 22 |
266989_at |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
0.602 |
0.022 |
| 23 |
256589_at |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
0.580 |
-0.020 |
| 24 |
260028_at |
AT1G29980
|
unknown |
0.566 |
-0.039 |
| 25 |
252607_at |
AT3G44990
|
AtXTH31, ATXTR8, XTH31, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 31, XTR8, xyloglucan endo-transglycosylase-related 8 |
0.549 |
-0.363 |
| 26 |
266168_at |
AT2G38870
|
[Serine protease inhibitor, potato inhibitor I-type family protein] |
0.525 |
-0.128 |
| 27 |
256159_at |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
0.475 |
0.009 |
| 28 |
254889_at |
AT4G11650
|
ATOSM34, osmotin 34, OSM34, osmotin 34 |
0.475 |
0.018 |
| 29 |
255186_at |
AT4G07750
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 2.9e-168 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
0.474 |
-0.048 |
| 30 |
257673_at |
AT3G20370
|
[TRAF-like family protein] |
0.462 |
-0.032 |
| 31 |
259383_at |
AT3G16470
|
JAL35, jacalin-related lectin 35, JR1, JASMONATE RESPONSIVE 1 |
0.460 |
0.034 |
| 32 |
250662_at |
AT5G07010
|
ATST2A, ARABIDOPSIS THALIANA SULFOTRANSFERASE 2A, ST2A, sulfotransferase 2A |
0.459 |
-0.013 |
| 33 |
249732_at |
AT5G24420
|
PGL5, 6-phosphogluconolactonase 5 |
0.430 |
0.062 |
| 34 |
263031_at |
AT1G24070
|
ATCSLA10, cellulose synthase-like A10, CSLA10, CELLULOSE SYNTHASE LIKE A10, CSLA10, cellulose synthase-like A10 |
0.424 |
-0.036 |
| 35 |
260560_at |
AT2G43590
|
[Chitinase family protein] |
0.404 |
-0.017 |
| 36 |
246550_at |
AT5G14920
|
GASA14, A-stimulated in Arabidopsis 14 |
0.396 |
0.022 |
| 37 |
263515_at |
AT2G21640
|
unknown |
0.372 |
-0.053 |
| 38 |
255285_at |
AT4G04630
|
unknown |
0.366 |
-0.046 |
| 39 |
256243_at |
AT3G12500
|
ATHCHIB, basic chitinase, B-CHI, CHI-B, HCHIB, basic chitinase, PR-3, PATHOGENESIS-RELATED 3, PR3, PATHOGENESIS-RELATED 3 |
0.364 |
0.017 |
| 40 |
256321_at |
AT1G55020
|
ATLOX1, ARABIDOPSIS LIPOXYGENASE 1, LOX1, lipoxygenase 1 |
0.362 |
-0.039 |
| 41 |
249073_at |
AT5G44020
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
0.352 |
-0.010 |
| 42 |
253073_at |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
0.349 |
-0.042 |
| 43 |
257809_at |
AT3G27060
|
ATTSO2, TSO2, TSO MEANING 'UGLY' IN CHINESE 2 |
0.345 |
-0.010 |
| 44 |
245393_at |
AT4G16260
|
[Glycosyl hydrolase superfamily protein] |
0.338 |
-0.014 |
| 45 |
250443_at |
AT5G10520
|
RBK1, ROP binding protein kinases 1 |
0.326 |
-0.013 |
| 46 |
266418_at |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
0.322 |
-0.069 |
| 47 |
254815_at |
AT4G12420
|
SKU5 |
0.321 |
-0.013 |
| 48 |
247327_at |
AT5G64120
|
AtPRX71, PRX71, peroxidase 71 |
0.321 |
0.214 |
| 49 |
254761_at |
AT4G13195
|
CLE44, CLAVATA3/ESR-RELATED 44 |
0.317 |
-0.026 |
| 50 |
266905_at |
AT2G34560
|
AtCCP1, CCP1, conserved in ciliated species and in the land plants 1 |
0.307 |
-0.051 |
| 51 |
257946_at |
AT3G21710
|
unknown |
0.305 |
-0.073 |
| 52 |
260541_at |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
0.301 |
0.030 |
| 53 |
246030_at |
AT5G21105
|
[Plant L-ascorbate oxidase] |
0.297 |
0.014 |
| 54 |
259375_at |
AT3G16370
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
0.295 |
0.001 |
| 55 |
253617_at |
AT4G30410
|
[sequence-specific DNA binding transcription factors] |
0.292 |
0.011 |
| 56 |
256441_at |
AT3G10940
|
LSF2, LIKE SEX4 2 |
0.285 |
-0.033 |
| 57 |
263693_at |
AT1G31200
|
ATPP2-A9, phloem protein 2-A9, PP2-A9, phloem protein 2-A9 |
0.279 |
0.040 |
| 58 |
247577_at |
AT5G61290
|
[Flavin-binding monooxygenase family protein] |
0.277 |
-0.013 |
| 59 |
258054_at |
AT3G16240
|
AQP1, ATTIP2;1, DELTA-TIP, delta tonoplast integral protein, DELTA-TIP1, TIP2;1 |
0.275 |
0.002 |
| 60 |
251545_at |
AT3G58810
|
ATMTP3, ARABIDOPSIS METAL TOLERANCE PROTEIN 3, ATMTPA2, MTP3, METAL TOLERANCE PROTEIN 3, MTPA2, metal tolerance protein A2 |
0.270 |
0.026 |
| 61 |
247069_at |
AT5G66920
|
sks17, SKU5 similar 17 |
0.261 |
-0.016 |
| 62 |
256913_at |
AT3G23870
|
unknown |
0.260 |
0.004 |
| 63 |
262637_at |
AT1G06640
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
0.253 |
0.009 |
| 64 |
255104_at |
AT4G08685
|
SAH7 |
0.247 |
-0.005 |
| 65 |
247162_at |
AT5G65730
|
XTH6, xyloglucan endotransglucosylase/hydrolase 6 |
0.246 |
-0.023 |
| 66 |
262414_at |
AT1G49430
|
LACS2, long-chain acyl-CoA synthetase 2, LRD2, LATERAL ROOT DEVELOPMENT 2 |
0.245 |
-0.040 |
| 67 |
254914_at |
AT4G11290
|
[Peroxidase superfamily protein] |
0.241 |
-0.028 |
| 68 |
252391_at |
AT3G47860
|
CHL, chloroplastic lipocalin |
0.241 |
-0.017 |
| 69 |
248622_at |
AT5G49360
|
ATBXL1, BETA-XYLOSIDASE 1, BXL1, beta-xylosidase 1 |
0.225 |
0.054 |
| 70 |
257206_at |
AT3G16530
|
[Legume lectin family protein] |
0.225 |
0.112 |
| 71 |
253627_at |
AT4G30650
|
[Low temperature and salt responsive protein family] |
0.223 |
-0.041 |
| 72 |
265149_at |
AT1G51400
|
[Photosystem II 5 kD protein] |
0.223 |
0.002 |
| 73 |
259384_at |
AT3G16450
|
JAL33, Jacalin-related lectin 33 |
0.221 |
-0.070 |
| 74 |
259431_at |
AT1G01620
|
PIP1;3, PLASMA MEMBRANE INTRINSIC PROTEIN 1;3, PIP1C, plasma membrane intrinsic protein 1C, TMP-B |
0.221 |
-0.015 |
| 75 |
253049_at |
AT4G37300
|
MEE59, maternal effect embryo arrest 59 |
0.217 |
-0.019 |
| 76 |
259366_at |
AT1G13280
|
AOC4, allene oxide cyclase 4 |
0.213 |
-0.010 |
| 77 |
265114_at |
AT1G62440
|
LRX2, leucine-rich repeat/extensin 2 |
0.208 |
0.039 |
| 78 |
248139_at |
AT5G54970
|
unknown |
0.208 |
-0.016 |
| 79 |
262694_at |
AT1G62790
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
0.208 |
0.051 |
| 80 |
256766_at |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-1.322 |
0.021 |
| 81 |
248322_at |
AT5G52760
|
[Copper transport protein family] |
-1.293 |
0.306 |
| 82 |
256593_at |
AT3G28510
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
-1.255 |
0.135 |
| 83 |
257382_at |
AT2G40750
|
ATWRKY54, WRKY DNA-BINDING PROTEIN 54, WRKY54, WRKY DNA-binding protein 54 |
-1.241 |
0.010 |
| 84 |
250942_at |
AT5G03350
|
[Legume lectin family protein] |
-1.238 |
0.060 |
| 85 |
259559_at |
AT1G21240
|
WAK3, wall associated kinase 3 |
-1.234 |
0.198 |
| 86 |
254271_at |
AT4G23150
|
CRK7, cysteine-rich RLK (RECEPTOR-like protein kinase) 7 |
-1.231 |
0.444 |
| 87 |
261221_at |
AT1G19960
|
unknown |
-1.224 |
0.002 |
| 88 |
248169_at |
AT5G54610
|
ANK, ankyrin, BDA1, bian da 2 (becoming big in Chinese) |
-1.206 |
0.181 |
| 89 |
265837_at |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-1.160 |
0.050 |
| 90 |
257100_at |
AT3G25010
|
AtRLP41, receptor like protein 41, RLP41, receptor like protein 41 |
-1.116 |
0.376 |
| 91 |
249754_at |
AT5G24530
|
DMR6, DOWNY MILDEW RESISTANT 6 |
-1.110 |
0.174 |
| 92 |
259385_at |
AT1G13470
|
unknown |
-1.082 |
0.388 |
| 93 |
260116_at |
AT1G33960
|
AIG1, AVRRPT2-INDUCED GENE 1 |
-1.073 |
0.529 |
| 94 |
245035_at |
AT2G26400
|
ARD, ACIREDUCTONE DIOXYGENASE, ARD3, acireductone dioxygenase 3, ATARD3, acireductone dioxygenase 3 |
-1.052 |
0.131 |
| 95 |
253730_at |
AT4G29480
|
[Mitochondrial ATP synthase subunit G protein] |
-1.021 |
-0.001 |
| 96 |
251705_at |
AT3G56400
|
ATWRKY70, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 70, WRKY70, WRKY DNA-binding protein 70 |
-1.005 |
0.047 |
| 97 |
251625_at |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
-0.976 |
0.394 |
| 98 |
255341_at |
AT4G04500
|
CRK37, cysteine-rich RLK (RECEPTOR-like protein kinase) 37 |
-0.950 |
0.380 |
| 99 |
265161_at |
AT1G30900
|
BP80-3;3, binding protein of 80 kDa 3;3, VSR3;3, VACUOLAR SORTING RECEPTOR 3;3, VSR6, VACUOLAR SORTING RECEPTOR 6 |
-0.946 |
0.230 |
| 100 |
267546_at |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
-0.944 |
0.281 |
| 101 |
260904_at |
AT1G02450
|
NIMIN-1, NIMIN1, NIM1-interacting 1 |
-0.939 |
0.106 |
| 102 |
254229_at |
AT4G23610
|
[Late embryogenesis abundant (LEA) hydroxyproline-rich glycoprotein family] |
-0.918 |
0.207 |
| 103 |
252549_at |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-0.916 |
0.224 |
| 104 |
254975_at |
AT4G10500
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-0.908 |
0.489 |
| 105 |
249890_at |
AT5G22570
|
ATWRKY38, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 38, WRKY38, WRKY DNA-binding protein 38 |
-0.898 |
0.166 |
| 106 |
259561_at |
AT1G21250
|
AtWAK1, PRO25, WAK1, cell wall-associated kinase 1 |
-0.895 |
0.104 |
| 107 |
248062_at |
AT5G55450
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.893 |
0.079 |
| 108 |
250445_at |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-0.876 |
0.301 |
| 109 |
261339_at |
AT1G35710
|
[Protein kinase family protein with leucine-rich repeat domain] |
-0.857 |
0.141 |
| 110 |
257591_at |
AT3G24900
|
AtRLP39, receptor like protein 39, RLP39, receptor like protein 39 |
-0.857 |
0.180 |
| 111 |
263539_at |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-0.857 |
0.324 |
| 112 |
245265_at |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.855 |
0.037 |
| 113 |
252060_at |
AT3G52430
|
ATPAD4, ARABIDOPSIS PHYTOALEXIN DEFICIENT 4, PAD4, PHYTOALEXIN DEFICIENT 4 |
-0.832 |
0.223 |
| 114 |
254231_at |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.816 |
0.231 |
| 115 |
254869_at |
AT4G11890
|
ARCK1, ABA- AND OSMOTIC-STRESS-INDUCIBLE RECEPTOR-LIKE CYTOSOLIC KINASE1, CRK45, cysteine-rich receptor-like protein kinase 45 |
-0.799 |
0.353 |
| 116 |
259925_at |
AT1G75040
|
PR-5, PR5, pathogenesis-related gene 5 |
-0.796 |
0.264 |
| 117 |
255595_at |
AT4G01700
|
[Chitinase family protein] |
-0.790 |
0.120 |
| 118 |
247684_at |
AT5G59670
|
[Leucine-rich repeat protein kinase family protein] |
-0.789 |
0.058 |
| 119 |
245329_at |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
-0.778 |
0.192 |
| 120 |
257185_at |
AT3G13100
|
ABCC7, ATP-binding cassette C7, ATMRP7, multidrug resistance-associated protein 7, MRP7, MRP7, multidrug resistance-associated protein 7 |
-0.752 |
0.224 |
| 121 |
245052_at |
AT2G26440
|
PME12, pectin methylesterase 12 |
-0.750 |
0.137 |
| 122 |
252345_at |
AT3G48640
|
unknown |
-0.742 |
0.241 |
| 123 |
249417_at |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
-0.742 |
0.305 |
| 124 |
263783_at |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
-0.737 |
0.203 |
| 125 |
250302_at |
AT5G11920
|
AtcwINV6, 6-&1-fructan exohydrolase, cwINV6, 6-&1-fructan exohydrolase |
-0.732 |
0.286 |
| 126 |
254255_at |
AT4G23220
|
CRK14, cysteine-rich RLK (RECEPTOR-like protein kinase) 14 |
-0.730 |
0.244 |
| 127 |
247314_at |
AT5G64000
|
ATSAL2, SAL2 |
-0.723 |
0.349 |
| 128 |
247602_at |
AT5G60900
|
RLK1, receptor-like protein kinase 1 |
-0.722 |
0.113 |
| 129 |
249743_at |
AT5G24540
|
BGLU31, beta glucosidase 31 |
-0.717 |
0.100 |
| 130 |
259550_at |
AT1G35230
|
AGP5, arabinogalactan protein 5 |
-0.716 |
0.291 |
| 131 |
252417_at |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
-0.709 |
0.268 |
| 132 |
252403_at |
AT3G48080
|
[alpha/beta-Hydrolases superfamily protein] |
-0.695 |
0.233 |
| 133 |
249918_at |
AT5G19240
|
[Glycoprotein membrane precursor GPI-anchored] |
-0.694 |
0.133 |
| 134 |
264434_at |
AT1G10340
|
[Ankyrin repeat family protein] |
-0.694 |
0.212 |
| 135 |
252131_at |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
-0.690 |
0.216 |
| 136 |
248321_at |
AT5G52740
|
[Copper transport protein family] |
-0.655 |
0.227 |
| 137 |
251970_at |
AT3G53150
|
UGT73D1, UDP-glucosyl transferase 73D1 |
-0.651 |
0.130 |
| 138 |
246368_at |
AT1G51890
|
[Leucine-rich repeat protein kinase family protein] |
-0.649 |
0.336 |
| 139 |
246405_at |
AT1G57630
|
[Toll-Interleukin-Resistance (TIR) domain family protein] |
-0.627 |
0.346 |
| 140 |
249777_at |
AT5G24210
|
[alpha/beta-Hydrolases superfamily protein] |
-0.618 |
0.089 |
| 141 |
254416_at |
AT4G21380
|
ARK3, receptor kinase 3, RK3, receptor kinase 3 |
-0.615 |
0.118 |
| 142 |
259805_at |
AT1G47890
|
AtRLP7, receptor like protein 7, RLP7, receptor like protein 7 |
-0.609 |
0.110 |
| 143 |
256366_at |
AT1G66880
|
[Protein kinase superfamily protein] |
-0.601 |
0.222 |
| 144 |
262666_at |
AT1G14080
|
ATFUT6, FUT6, fucosyltransferase 6 |
-0.598 |
-0.006 |
| 145 |
261443_at |
AT1G28480
|
GRX480, roxy19 |
-0.591 |
0.140 |
| 146 |
259560_at |
AT1G21270
|
WAK2, wall-associated kinase 2 |
-0.584 |
0.050 |
| 147 |
266761_at |
AT2G47130
|
AtSDR3, SDR3, short-chain dehydrogenase/reductase 2 |
-0.579 |
0.217 |
| 148 |
265723_at |
AT2G32140
|
[transmembrane receptors] |
-0.575 |
0.203 |
| 149 |
263852_at |
AT2G04450
|
ATNUDT6, nudix hydrolase homolog 6, ATNUDX6, Arabidopsis thaliana nucleoside diphosphate linked to some moiety X 6, NUDT6, nudix hydrolase homolog 6, NUDX6, nucleoside diphosphates linked to some moiety X 6 |
-0.575 |
0.195 |
| 150 |
259428_at |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
-0.575 |
0.194 |
| 151 |
266782_at |
AT2G29120
|
ATGLR2.7, glutamate receptor 2.7, GLR2.7, GLUTAMATE RECEPTOR 2.7, GLR2.7, glutamate receptor 2.7 |
-0.570 |
0.291 |
| 152 |
267436_at |
AT2G19190
|
FRK1, FLG22-induced receptor-like kinase 1 |
-0.565 |
0.028 |
| 153 |
264223_s_at |
AT3G16030
|
CES101, CALLUS EXPRESSION OF RBCS 101, RFO3, RESISTANCE TO FUSARIUM OXYSPORUM 3 |
-0.563 |
0.166 |
| 154 |
246988_at |
AT5G67340
|
[ARM repeat superfamily protein] |
-0.552 |
0.224 |
| 155 |
262507_at |
AT1G11330
|
[S-locus lectin protein kinase family protein] |
-0.549 |
0.045 |
| 156 |
254660_at |
AT4G18250
|
[receptor serine/threonine kinase, putative] |
-0.543 |
0.385 |
| 157 |
252373_at |
AT3G48090
|
ATEDS1, EDS1, enhanced disease susceptibility 1 |
-0.534 |
0.108 |