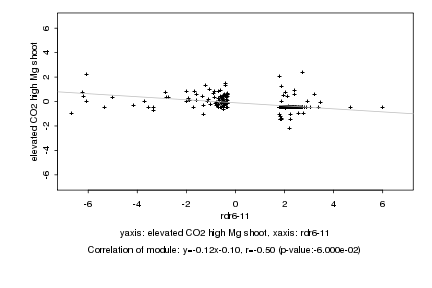
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
rdr6-11 |
Log2 signal ratio
elevated CO2 high Mg shoot |
| 1 |
AT2G21360 |
AT2G21360
|
[tRNA-Pro (anticodon: CGG)] |
5.959 |
-0.467 |
| 2 |
AT4G21370 |
AT4G21370
|
pseudogene |
4.674 |
-0.467 |
| 3 |
AT4G33100 |
AT4G33100
|
unknown |
3.445 |
-0.039 |
| 4 |
AT2G15340 |
AT2G15340
|
[glycine-rich protein] |
3.383 |
-0.467 |
| 5 |
AT5G40020 |
AT5G40020
|
[Pathogenesis-related thaumatin superfamily protein] |
3.200 |
0.620 |
| 6 |
AT2G04990 |
AT2G04990
|
unknown |
3.056 |
-0.467 |
| 7 |
AT3G44096 |
AT3G44096
|
[non-LTR retrotransposon family (LINE), has a 4.8e-23 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
2.914 |
0.055 |
| 8 |
AT3G42600 |
AT3G42600
|
unknown |
2.859 |
-0.467 |
| 9 |
AT3G43640 |
AT3G43640
|
[copia-like retrotransposon family, has a 1.4e-25 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
2.779 |
-0.467 |
| 10 |
AT1G62045 |
AT1G62045
|
unknown |
2.743 |
-0.923 |
| 11 |
AT3G26610 |
AT3G26610
|
[Pectin lyase-like superfamily protein] |
2.702 |
-0.467 |
| 12 |
AT1G43660 |
AT1G43660
|
unknown |
2.697 |
-0.467 |
| 13 |
AT1G77640 |
AT1G77640
|
[Integrase-type DNA-binding superfamily protein] |
2.692 |
2.447 |
| 14 |
AT4G04635 |
AT4G04635
|
unknown |
2.692 |
-0.467 |
| 15 |
AT5G37080 |
AT5G37080
|
unknown |
2.665 |
-0.467 |
| 16 |
AT1G64340 |
AT1G64340
|
unknown |
2.632 |
-0.467 |
| 17 |
AT3G28695 |
AT3G28695
|
[tRNA-Ala (anticodon: AGC)] |
2.603 |
-0.467 |
| 18 |
AT4G23387 |
AT4G23387
|
MIR845A, microRNA845A |
2.547 |
-0.467 |
| 19 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
2.544 |
-0.925 |
| 20 |
AT1G35940 |
AT1G35940
|
unknown |
2.534 |
-0.467 |
| 21 |
AT4G06549 |
AT4G06549
|
[pseudogene] |
2.515 |
-0.467 |
| 22 |
AT5G29058 |
AT5G29058
|
[pseudogene] |
2.482 |
-0.467 |
| 23 |
AT2G07300 |
AT2G07300
|
unknown |
2.481 |
-0.467 |
| 24 |
AT1G43763 |
AT1G43763
|
[copia-like retrotransposon family, has a 1.9e-96 P-value blast match to GB:AAB82754 retrofit (TY1_Copia-element) (Oryza longistaminata)] |
2.433 |
-0.467 |
| 25 |
AT4G06496 |
AT4G06496
|
[pseudogene] |
2.429 |
-0.467 |
| 26 |
AT5G35348 |
AT5G35348
|
[pseudogene] |
2.424 |
-0.467 |
| 27 |
AT5G33990 |
AT5G33990
|
[gypsy-like retrotransposon family (Athila), has a 5.0e-73 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
2.393 |
-0.467 |
| 28 |
AT4G25810 |
AT4G25810
|
XTH23, xyloglucan endotransglucosylase/hydrolase 23, XTR6, xyloglucan endotransglycosylase 6 |
2.389 |
0.921 |
| 29 |
AT3G50350 |
AT3G50350
|
unknown |
2.387 |
0.606 |
| 30 |
AT4G06522 |
AT4G06522
|
[non-LTR retrotransposon family (LINE), has a 1.2e-22 P-value blast match to GB:NP_038603 L1 repeat, Tf subfamily, member 23 (LINE-element) (Mus musculus)] |
2.373 |
-0.467 |
| 31 |
AT5G35475 |
AT5G35475
|
[General transcription factor 2-related zinc finger protein] |
2.363 |
-0.467 |
| 32 |
AT2G12870 |
AT2G12870
|
[gypsy-like retrotransposon family, has a 3.8e-07 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
2.356 |
-0.467 |
| 33 |
AT5G35695 |
AT5G35695
|
unknown |
2.336 |
-0.467 |
| 34 |
AT5G28527 |
AT5G28527
|
[pseudogene] |
2.334 |
-0.467 |
| 35 |
AT3G42723 |
AT3G42723
|
[aminoacyl-tRNA ligases] |
2.306 |
-0.467 |
| 36 |
AT5G35640 |
AT5G35640
|
[Putative endonuclease or glycosyl hydrolase] |
2.298 |
-0.467 |
| 37 |
AT1G56675 |
AT1G56675
|
[copia-like retrotransposon family, has a 1.7e-196 P-value blast match to GB:AAC02666 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
2.286 |
-0.467 |
| 38 |
AT5G28667 |
AT5G28667
|
[pseudogene] |
2.286 |
-0.467 |
| 39 |
AT3G33115 |
AT3G33115
|
[gypsy-like retrotransposon family, has a 2.9e-169 P-value blast match to GB:AAD11615 prpol (gypsy_Ty3-element) (Zea mays)] |
2.280 |
-0.467 |
| 40 |
AT3G45840 |
AT3G45840
|
[Cysteine/Histidine-rich C1 domain family protein] |
2.253 |
-0.467 |
| 41 |
AT5G28930 |
AT5G28930
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT2G07240.1)] |
2.248 |
-0.467 |
| 42 |
AT5G35965 |
AT5G35965
|
[copia-like retrotransposon family, has a 5.9e-244 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
2.240 |
-1.006 |
| 43 |
AT1G35990 |
AT1G35990
|
[non-LTR retrotransposon family (LINE), has a 1.2e-23 P-value blast match to GB:NP_038602 L1 repeat, Tf subfamily, member 18 (LINE-element) (Mus musculus)] |
2.240 |
-0.467 |
| 44 |
AT4G07519 |
AT4G07519
|
[pseudogene] |
2.223 |
-0.467 |
| 45 |
AT3G43260 |
AT3G43260
|
unknown |
2.221 |
-0.467 |
| 46 |
AT4G18410 |
AT4G18410
|
[Mutator-like transposase family, has a 8.7e-30 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
2.213 |
-1.398 |
| 47 |
AT3G33073 |
AT3G33073
|
transposable element gene |
2.213 |
-0.467 |
| 48 |
AT5G35923 |
AT5G35923
|
[gypsy-like retrotransposon family (Athila), has a 2.6e-33 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
2.200 |
-0.467 |
| 49 |
AT1G37000 |
AT1G37000
|
[Beta-galactosidase related protein] |
2.183 |
-0.467 |
| 50 |
AT3G42870 |
AT3G42870
|
unknown |
2.179 |
-0.467 |
| 51 |
AT5G36300 |
AT5G36300
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
2.175 |
-0.467 |
| 52 |
AT3G43126 |
AT3G43126
|
[CACTA-like transposase family (En/Spm), has a 6.3e-145 P-value blast match to GB:AAD55677 putative transposase protein (CACTA-element) transposon=Shooter (Zea mays)] |
2.174 |
-0.467 |
| 53 |
AT5G29591 |
AT5G29591
|
[Mutator-like transposase family, has a 1.4e-14 P-value blast match to Q9XE24 /118-277 Pfam PF03108 MuDR family transposase (MuDr-element domain)] |
2.172 |
-0.467 |
| 54 |
AT3G29033 |
AT3G29033
|
[glycine-rich protein] |
2.170 |
-0.467 |
| 55 |
AT5G32518 |
AT5G32518
|
[gypsy-like retrotransposon family, has a 1.5e-246 P-value blast match to GB:CAA73042 polyprotein (Gypsy_Ty3-element) (Ananas comosus)] |
2.170 |
-0.467 |
| 56 |
AT3G53065 |
AT3G53065
|
[D-galactoside/L-rhamnose binding SUEL lectin protein] |
2.167 |
-2.175 |
| 57 |
AT5G35720 |
AT5G35720
|
[non-LTR retrotransposon family (LINE), has a 6.9e-12 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
2.133 |
-0.467 |
| 58 |
AT5G66070 |
AT5G66070
|
[RING/U-box superfamily protein] |
2.132 |
-0.334 |
| 59 |
AT2G14390 |
AT2G14390
|
unknown |
2.130 |
-0.467 |
| 60 |
AT4G06570 |
AT4G06570
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.3e-26 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
2.118 |
-0.467 |
| 61 |
AT2G12340 |
AT2G12340
|
[non-LTR retrotransposon family (LINE), has a 2.6e-31 P-value blast match to GB:NP_038605 L1 repeat, Tf subfamily, member 30 (LINE-element) (Mus musculus)] |
2.113 |
-0.467 |
| 62 |
AT1G62670 |
AT1G62670
|
RPF2, rna processing factor 2 |
2.112 |
0.415 |
| 63 |
AT4G03835 |
AT4G03835
|
[gypsy-like retrotransposon family, has a 6.1e-78 P-value blast match to GB:BAA84458 GAG-POL precursor (gypsy_Ty3-element) (Oryza sativa)gi|5902445|dbj|BAA84458.1| GAG-POL precursor (Oryza sativa (japonica cultivar-group)) (RIRE2) (Gypsy_Ty3-family)] |
2.104 |
-0.467 |
| 64 |
AT3G44230 |
AT3G44230
|
unknown |
2.102 |
-0.467 |
| 65 |
AT4G06556 |
AT4G06556
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 3.3e-152 P-value blast match to gb|AAG52024.1|AC022456_5 Tam1-homologous transposon protein TNP2, putative] |
2.076 |
-0.467 |
| 66 |
AT5G03400 |
AT5G03400
|
unknown |
2.075 |
-0.467 |
| 67 |
AT3G30213 |
AT3G30213
|
[gypsy-like retrotransposon family (Athila), has a 3.2e-53 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
2.066 |
-0.467 |
| 68 |
AT5G35526 |
AT5G35526
|
unknown |
2.062 |
-0.467 |
| 69 |
AT4G06497 |
AT4G06497
|
[non-LTR retrotransposon family (LINE), has a 5.0e-48 P-value blast match to GB:NP_038607 L1 repeat, Tf subfamily, member 9 (LINE-element) (Mus musculus)] |
2.057 |
-0.467 |
| 70 |
AT3G33124 |
AT3G33124
|
[gypsy-like retrotransposon family (Athila), has a 6.5e-171 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
2.051 |
-0.467 |
| 71 |
AT5G28821 |
AT5G28821
|
unknown |
2.044 |
-0.467 |
| 72 |
AT3G31402 |
AT3G31402
|
[General transcription factor 2-related zinc finger protein] |
2.039 |
-0.467 |
| 73 |
AT4G06597 |
AT4G06597
|
[pseudogene] |
2.039 |
-0.556 |
| 74 |
AT4G06595 |
AT4G06595
|
[copia-like retrotransposon family, has a 5.3e-49 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
2.032 |
-0.467 |
| 75 |
AT2G06110 |
AT2G06110
|
[gypsy-like retrotransposon family (Athila), has a 6.4e-72 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
2.030 |
-0.467 |
| 76 |
AT3G30560 |
AT3G30560
|
[similar to AT hook motif-containing protein-related [Arabidopsis thaliana] (TAIR:AT1G35940.1)] |
2.029 |
-0.467 |
| 77 |
AT3G33079 |
AT3G33079
|
[gypsy-like retrotransposon family (Athila), has a 1.7e-203 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
2.016 |
-0.467 |
| 78 |
AT1G67105 |
AT1G67105
|
other RNA |
2.014 |
0.782 |
| 79 |
AT1G21202 |
AT1G21202
|
MIR781A, microRNA781A |
2.012 |
-0.467 |
| 80 |
AT1G41790 |
AT1G41790
|
[gypsy-like retrotransposon family, has a 0.00017 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
1.996 |
-0.467 |
| 81 |
AT4G10980 |
AT4G10980
|
[copia-like retrotransposon family, has a 2.7e-44 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
1.991 |
-0.467 |
| 82 |
AT5G31496 |
AT5G31496
|
[gypsy-like retrotransposon family, has a 8.1e-10 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
1.991 |
-0.467 |
| 83 |
AT1G41810 |
AT1G41810
|
unknown |
1.990 |
-0.467 |
| 84 |
AT4G07425 |
AT4G07425
|
[CACTA-like transposase family (Tnp1/En/Spm), has a 2.5e-22 P-value blast match to ref|NP_189784.1| TNP1-related protein (Arabidopsis thaliana) (CACTA-element)] |
1.989 |
-0.467 |
| 85 |
AT5G32590 |
AT5G32590
|
[myosin heavy chain-related] |
1.978 |
-0.467 |
| 86 |
AT5G28970 |
AT5G28970
|
[similar to Ulp1 protease family protein [Arabidopsis thaliana] (TAIR:AT1G35110.1)] |
1.974 |
-0.467 |
| 87 |
AT1G42605 |
AT1G42605
|
[gypsy-like retrotransposon family, has a 5.9e-318 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
1.960 |
-0.467 |
| 88 |
AT5G29337 |
AT5G29337
|
[gypsy-like retrotransposon family (Athila), has a 2.7e-77 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.952 |
-0.467 |
| 89 |
AT4G14504 |
AT4G14504
|
MIR852A, microRNA852A |
1.952 |
-0.467 |
| 90 |
AT3G30703 |
AT3G30703
|
[gypsy-like retrotransposon family, has a 6.1e-20 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
1.941 |
-0.467 |
| 91 |
AT1G47360 |
AT1G47360
|
[copia-like retrotransposon family, has a 1.2e-307 P-value blast match to GB:CAA31653 polyprotein (Ty1_Copia-element) (Arabidopsis thaliana)] |
1.940 |
-0.467 |
| 92 |
AT4G06488 |
AT4G06488
|
[gypsy-like retrotransposon family, has a 3.5e-167 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
1.939 |
-0.467 |
| 93 |
AT2G11168 |
AT2G11168
|
[pseudogene] |
1.939 |
-0.467 |
| 94 |
AT4G21940 |
AT4G21940
|
CPK15, calcium-dependent protein kinase 15 |
1.937 |
-0.426 |
| 95 |
AT5G30218 |
AT5G30218
|
[gypsy-like retrotransposon family (Athila), has a 3.3e-30 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.937 |
-0.467 |
| 96 |
AT1G35617 |
AT1G35617
|
unknown |
1.934 |
-0.467 |
| 97 |
AT4G07733 |
AT4G07733
|
[gypsy-like retrotransposon family (Athila), has a 2.5e-37 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.929 |
-0.467 |
| 98 |
AT4G03690 |
AT4G03690
|
unknown |
1.928 |
-0.467 |
| 99 |
AT2G10320 |
AT2G10320
|
[gypsy-like retrotransposon family (Athila), has a 2.3e-75 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.924 |
-0.467 |
| 100 |
AT3G44115 |
AT3G44115
|
unknown |
1.923 |
-0.467 |
| 101 |
AT1G48840 |
AT1G48840
|
unknown |
1.922 |
0.499 |
| 102 |
AT1G43330 |
AT1G43330
|
[Homeodomain-like superfamily protein] |
1.919 |
-0.467 |
| 103 |
AT4G07528 |
AT4G07528
|
[gypsy-like retrotransposon family, has a 1.6e-141 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
1.912 |
-0.467 |
| 104 |
AT5G36080 |
AT5G36080
|
unknown |
1.906 |
-0.467 |
| 105 |
AT4G03670 |
AT4G03670
|
unknown |
1.903 |
-0.467 |
| 106 |
AT3G42445 |
AT3G42445
|
[Athila retroelement ORF2 -related] |
1.901 |
-0.467 |
| 107 |
AT2G13710 |
AT2G13710
|
[Mariner-like transposase family, has a 3.3e-55 P-value blast match to GB:AAC28384 mariner transposase (Mariner_TC1-element) (Glycine max)] |
1.901 |
-0.467 |
| 108 |
AT5G28535 |
AT5G28535
|
[Mariner-like transposase family, has a 1.2e-97 P-value blast match to GB:AAC28384 mariner transposase (Mariner_TC1-element) (Glycine max)] |
1.897 |
-0.467 |
| 109 |
AT2G13040 |
AT2G13040
|
[pseudogene] |
1.892 |
-0.467 |
| 110 |
AT3G32091 |
AT3G32091
|
[gypsy-like retrotransposon family, has a 1.3e-109 P-value blast match to F1M23 reverse transcriptase (from Dan Voytas http://www.public.iastate.edu/~voytas) (Gypsy_Ty3-family)] |
1.883 |
-0.467 |
| 111 |
AT5G27480 |
AT5G27480
|
[pseudogene] |
1.883 |
-0.467 |
| 112 |
AT4G04550 |
AT4G04550
|
[copia-like retrotransposon family, has a 1.9e-10 P-value blast match to gb|AAG52950.1| putative envelope protein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
1.881 |
-0.467 |
| 113 |
AT4G06639 |
AT4G06639
|
unknown |
1.877 |
-0.467 |
| 114 |
AT4G05632 |
AT4G05632
|
unknown |
1.876 |
-0.467 |
| 115 |
AT1G36230 |
AT1G36230
|
unknown |
1.876 |
-0.467 |
| 116 |
AT5G28820 |
AT5G28820
|
unknown |
1.876 |
-0.467 |
| 117 |
AT5G32515 |
AT5G32515
|
[pseudogene] |
1.876 |
-0.467 |
| 118 |
AT4G04310 |
AT4G04310
|
[pseudogene] |
1.871 |
0.055 |
| 119 |
AT5G35470 |
AT5G35470
|
unknown |
1.870 |
-0.467 |
| 120 |
AT1G40103 |
AT1G40103
|
[pseudogene] |
1.870 |
-0.467 |
| 121 |
AT1G35614 |
AT1G35614
|
unknown |
1.870 |
-0.467 |
| 122 |
AT3G10930 |
AT3G10930
|
unknown |
1.870 |
-1.329 |
| 123 |
AT4G30110 |
AT4G30110
|
ATHMA2, ARABIDOPSIS HEAVY METAL ATPASE 2, HMA2, heavy metal atpase 2 |
1.863 |
1.251 |
| 124 |
AT5G36015 |
AT5G36015
|
[pseudogene] |
1.863 |
-0.467 |
| 125 |
AT1G31358 |
AT1G31358
|
MIR404, microRNA404 |
1.862 |
-0.467 |
| 126 |
AT2G10833 |
AT2G10833
|
[pseudogene] |
1.859 |
-0.467 |
| 127 |
AT5G57560 |
AT5G57560
|
TCH4, Touch 4, XTH22, xyloglucan endotransglucosylase/hydrolase 22 |
1.847 |
-1.468 |
| 128 |
AT3G44170 |
AT3G44170
|
unknown |
1.834 |
-0.467 |
| 129 |
AT3G48057 |
AT3G48057
|
MIR843A, microRNA843A |
1.830 |
-0.467 |
| 130 |
AT4G04230 |
AT4G04230
|
[gypsy-like retrotransposon family, has a 1.3e-95 P-value blast match to T27D20 reverse transcriptase (from Dan Voytas http://www.public.iastate.edu/~voytas) (Gypsy_Ty3-family)] |
1.828 |
-0.467 |
| 131 |
AT4G06479 |
AT4G06479
|
[zinc ion binding] |
1.828 |
-0.467 |
| 132 |
AT5G28141 |
AT5G28141
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 5.0e-54 P-value blast match to GB:CAA38906 Tam3-transposase (hAT-element) (Antirrhinum majus)] |
1.828 |
-0.467 |
| 133 |
AT1G66640 |
AT1G66640
|
[RNI-like superfamily protein] |
1.828 |
-0.467 |
| 134 |
AT4G19240 |
AT4G19240
|
unknown |
1.828 |
-0.467 |
| 135 |
AT4G07490 |
AT4G07490
|
[transposon protein -related] |
1.828 |
-0.467 |
| 136 |
AT2G16610 |
AT2G16610
|
[CACTA-like transposase family (En/Spm), has a 6.1e-89 P-value blast match to GB:BAA20532 ORF of transposon Tdc1 (CACTA-element) (Daucus carota)] |
1.828 |
-0.467 |
| 137 |
AT2G10253 |
AT2G10253
|
[pseudogene] |
1.828 |
-1.398 |
| 138 |
AT4G06564 |
AT4G06564
|
[gypsy-like retrotransposon family (Athila), has a 2.5e-114 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.828 |
-0.467 |
| 139 |
AT4G05616 |
AT4G05616
|
unknown |
1.828 |
-0.467 |
| 140 |
AT5G32520 |
AT5G32520
|
[pseudogene] |
1.810 |
-0.467 |
| 141 |
AT3G30668 |
AT3G30668
|
[gypsy-like retrotransposon family (Athila), has a 6.6e-229 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.807 |
-0.467 |
| 142 |
AT4G06682 |
AT4G06682
|
[copia-like retrotransposon family, has a 1.2e-196 P-value blast match to gb|AAG52949.1| gag/pol polyprotein (Endovir1-1) (Arabidopsis thaliana) (Ty1_Copia-family)] |
1.801 |
-0.467 |
| 143 |
AT2G24070 |
AT2G24070
|
QWRF4, QWRF domain containing 4 |
1.801 |
-1.398 |
| 144 |
AT5G46195 |
AT5G46195
|
[hAT-like transposase family (hobo/Ac/Tam3), has a 8.8e-38 P-value blast match to GB:AAD24567 transposase Tag2 (hAT-element) (Arabidopsis thaliana)] |
1.798 |
-0.467 |
| 145 |
AT1G28370 |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
1.796 |
-1.178 |
| 146 |
AT2G11430 |
AT2G11430
|
[gypsy-like retrotransposon family, has a 0. P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
1.796 |
-0.467 |
| 147 |
AT5G29037 |
AT5G29037
|
[gypsy-like retrotransposon family, has a 2.2e-209 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
1.794 |
-0.467 |
| 148 |
AT5G34845 |
AT5G34845
|
[pseudogene] |
1.793 |
-0.467 |
| 149 |
AT3G42560 |
AT3G42560
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
1.793 |
-0.467 |
| 150 |
AT1G42080 |
AT1G42080
|
unknown |
1.792 |
-0.467 |
| 151 |
AT4G05505 |
AT4G05505
|
[CACTA-like transposase family (Ptta/En/Spm), has a 1.2e-35 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
1.789 |
-0.467 |
| 152 |
AT5G33422 |
AT5G33422
|
[CACTA-like transposase family (Tnp2/En/Spm), has a 3.5e-121 P-value blast match to GB:CAA40555 TNP2 (CACTA-element) (Antirrhinum majus)] |
1.785 |
-0.467 |
| 153 |
AT3G30650 |
AT3G30650
|
unknown |
1.784 |
-0.467 |
| 154 |
AT3G28958 |
AT3G28958
|
[Cupredoxin superfamily protein] |
1.784 |
-1.006 |
| 155 |
AT5G30406 |
AT5G30406
|
[gypsy-like retrotransposon family (Athila), has a 8.8e-131 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.779 |
-0.467 |
| 156 |
AT4G12915 |
AT4G12915
|
[gypsy-like retrotransposon family (Athila), has a 1.4e-20 P-value blast match to GB:CAA57397 Athila ORF 1 (Arabidopsis thaliana)] |
1.778 |
-0.467 |
| 157 |
AT3G29727 |
AT3G29727
|
[non-LTR retrotransposon family (LINE), has a 5.9e-09 P-value blast match to GB:AAA67727 reverse transcriptase (LINE-element) (Mus musculus)] |
1.778 |
-0.467 |
| 158 |
AT5G35926 |
AT5G35926
|
[Protein with RNI-like/FBD-like domains] |
1.778 |
2.067 |
| 159 |
AT1G32040 |
AT1G32040
|
unknown |
1.776 |
-0.467 |
| 160 |
AT4G06594 |
AT4G06594
|
[gypsy-like retrotransposon family, has a 5.1e-215 P-value blast match to GB:AAD27547 polyprotein (Gypsy_Ty3-element) (Oryza sativa subsp. indica)] |
1.773 |
-0.467 |
| 161 |
AT3G33009 |
AT3G33009
|
[pseudogene] |
1.770 |
-0.467 |
| 162 |
AT3G33163 |
AT3G33163
|
[pseudogene] |
1.769 |
-0.467 |
| 163 |
ATCG01160 |
ATCG01160
|
RRN5S, ribosomal RNA5S |
-6.710 |
-0.901 |
| 164 |
AT2G27400 |
AT2G27400
|
TAS1A, trans-acting siRNA1A |
-6.262 |
0.819 |
| 165 |
AT2G39681 |
AT2G39681
|
TAS2, trans-acting siRNA2 |
-6.216 |
0.468 |
| 166 |
AT1G50055 |
AT1G50055
|
TAS1B, trans-acting siRNA1B |
-6.072 |
0.030 |
| 167 |
AT2G39675 |
AT2G39675
|
TAS1C, trans-acting siRNA1C |
-6.071 |
2.227 |
| 168 |
AT3G25795 |
AT3G25795
|
TAS4, trans acting siRNA 4 |
-5.340 |
-0.467 |
| 169 |
AT5G42980 |
AT5G42980
|
ATH3, thioredoxin H-type 3, ATTRX3, thioredoxin 3, ATTRXH3, TRX3, thioredoxin 3, TRXH3, THIOREDOXIN H3 |
-5.035 |
0.333 |
| 170 |
AT1G63150 |
AT1G63150
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-4.157 |
-0.252 |
| 171 |
AT3G17185 |
AT3G17185
|
ATTAS3, TAS3, trans-acting siRNA3, TASIR-ARF |
-3.720 |
0.060 |
| 172 |
AT5G49615 |
AT5G49615
|
TAS3b, trans-acting siRNA3B |
-3.551 |
-0.467 |
| 173 |
AT1G63630 |
AT1G63630
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-3.378 |
-0.467 |
| 174 |
AT1G63080 |
AT1G63080
|
[Pentatricopeptide repeat (PPR) superfamily protein] |
-3.353 |
-0.694 |
| 175 |
AT5G38850 |
AT5G38850
|
[Disease resistance protein (TIR-NBS-LRR class)] |
-2.864 |
0.759 |
| 176 |
AT3G49500 |
AT3G49500
|
RDR6, RNA-dependent RNA polymerase 6, SDE1, SILENCING DEFECTIVE 1, SGS2, SUPPRESSOR OF GENE SILENCING 2 |
-2.824 |
0.360 |
| 177 |
AT1G63130 |
AT1G63130
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-2.753 |
0.376 |
| 178 |
AT2G01020 |
AT2G01020
|
[5.8SrRNA] |
-2.016 |
0.897 |
| 179 |
AT4G16590 |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
-2.005 |
0.065 |
| 180 |
AT2G46070 |
AT2G46070
|
ATMPK12, MAPK12, MPK12, mitogen-activated protein kinase 12 |
-1.924 |
0.272 |
| 181 |
AT1G62930 |
AT1G62930
|
RPF3, RNA processing factor 3 |
-1.874 |
0.133 |
| 182 |
AT5G57735 |
AT5G57735
|
TASIR-ARF |
-1.717 |
-0.467 |
| 183 |
AT1G03530 |
AT1G03530
|
ATNAF1, NAF1, nuclear assembly factor 1 |
-1.669 |
0.897 |
| 184 |
AT5G43740 |
AT5G43740
|
[Disease resistance protein (CC-NBS-LRR class) family] |
-1.613 |
0.122 |
| 185 |
AT4G15210 |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.598 |
0.600 |
| 186 |
AT2G01010 |
AT2G01010
|
[18SrRNA] |
-1.369 |
0.453 |
| 187 |
AT2G37240 |
AT2G37240
|
[Thioredoxin superfamily protein] |
-1.312 |
-0.247 |
| 188 |
AT5G25090 |
AT5G25090
|
AtENODL13, ENODL13, early nodulin-like protein 13 |
-1.312 |
-1.006 |
| 189 |
AT3G55500 |
AT3G55500
|
ATEXP16, ATEXPA16, expansin A16, ATHEXP ALPHA 1.7, EXP16, EXPANSIN 16, EXPA16, expansin A16 |
-1.243 |
1.360 |
| 190 |
AT1G14250 |
AT1G14250
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-1.140 |
0.009 |
| 191 |
AT3G19810 |
AT3G19810
|
unknown |
-1.111 |
0.241 |
| 192 |
AT1G11160 |
AT1G11160
|
[Transducin/WD40 repeat-like superfamily protein] |
-1.073 |
0.989 |
| 193 |
AT4G01080 |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
-1.053 |
-0.190 |
| 194 |
AT3G10690 |
AT3G10690
|
GYRA, DNA GYRASE A |
-0.916 |
0.723 |
| 195 |
AT3G17170 |
AT3G17170
|
RFC3, REGULATOR OF FATTY-ACID COMPOSITION 3 |
-0.875 |
0.900 |
| 196 |
AT3G11660 |
AT3G11660
|
NHL1, NDR1/HIN1-like 1 |
-0.860 |
0.404 |
| 197 |
AT3G18390 |
AT3G18390
|
EMB1865, embryo defective 1865 |
-0.815 |
-0.136 |
| 198 |
AT5G49940 |
AT5G49940
|
ATCNFU2, CHLOROPLAST-LOCALIZED NIFU-LIKE PROTEIN 2, NFU2, NIFU-like protein 2 |
-0.793 |
-0.037 |
| 199 |
AT1G69530 |
AT1G69530
|
AT-EXP1, ATEXP1, ATEXPA1, expansin A1, ATHEXP ALPHA 1.2, EXP1, EXPANSIN 1, EXPA1, expansin A1 |
-0.788 |
-0.288 |
| 200 |
AT5G17710 |
AT5G17710
|
EMB1241, embryo defective 1241 |
-0.737 |
-0.258 |
| 201 |
AT3G16250 |
AT3G16250
|
NDF4, NDH-dependent cyclic electron flow 1, PnsB3, Photosynthetic NDH subcomplex B 3 |
-0.719 |
0.291 |
| 202 |
AT3G04550 |
AT3G04550
|
unknown |
-0.706 |
-0.179 |
| 203 |
AT1G14370 |
AT1G14370
|
APK2A, protein kinase 2A, Kin1, kinase 1, PBL2, PBS1-like 2 |
-0.704 |
0.829 |
| 204 |
AT4G34035 |
AT4G34035
|
[tRNA-Arg (anticodon: TCT)] |
-0.698 |
-0.467 |
| 205 |
AT1G51060 |
AT1G51060
|
HTA10, histone H2A 10 |
-0.686 |
0.130 |
| 206 |
AT5G34790 |
AT5G34790
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.9e-11 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.641 |
0.946 |
| 207 |
AT2G22280 |
AT2G22280
|
[tRNA-Arg (anticodon: ACG)] |
-0.625 |
-0.467 |
| 208 |
AT3G47650 |
AT3G47650
|
[DnaJ/Hsp40 cysteine-rich domain superfamily protein] |
-0.612 |
0.418 |
| 209 |
AT3G02715 |
AT3G02715
|
[tRNA-Ser (anticodon: AGA)] |
-0.605 |
-0.467 |
| 210 |
ATCG00370 |
ATCG00370
|
TRNS.3 |
-0.602 |
-0.004 |
| 211 |
AT5G15970 |
AT5G15970
|
AtCor6.6, COR6.6, COLD-RESPONSIVE 6.6, KIN2 |
-0.591 |
0.343 |
| 212 |
AT5G13630 |
AT5G13630
|
ABAR, ABA-BINDING PROTEIN, CCH, CONDITIONAL CHLORINA, CCH1, CHLH, H SUBUNIT OF MG-CHELATASE, GUN5, GENOMES UNCOUPLED 5 |
-0.586 |
-0.120 |
| 213 |
AT3G26570 |
AT3G26570
|
ORF02, PHT2;1, phosphate transporter 2;1 |
-0.567 |
0.252 |
| 214 |
AT4G39800 |
AT4G39800
|
ATIPS1, INOSITOL 3-PHOSPHATE SYNTHASE 1, ATMIPS1, MYO-INOSITOL-1-PHOSPHATE SYNTHASE 1, MI-1-P SYNTHASE, MIPS1, D-myo-Inositol 3-Phosphate Synthase 1, MIPS1, myo-inositol-1-phosphate synthase 1 |
-0.566 |
0.230 |
| 215 |
AT5G49910 |
AT5G49910
|
cpHsc70-2, chloroplast heat shock protein 70-2, HSC70-7, HEAT SHOCK PROTEIN 70-7 |
-0.546 |
0.323 |
| 216 |
AT2G26080 |
AT2G26080
|
AtGLDP2, glycine decarboxylase P-protein 2, GLDP2, glycine decarboxylase P-protein 2 |
-0.529 |
-0.380 |
| 217 |
AT1G09780 |
AT1G09780
|
iPGAM1, 2,3-biphosphoglycerate-independent phosphoglycerate mutase 1 |
-0.520 |
0.037 |
| 218 |
AT5G56030 |
AT5G56030
|
AtHsp90.2, HEAT SHOCK PROTEIN 90.2, ERD8, EARLY-RESPONSIVE TO DEHYDRATION 8, HSP81-2, heat shock protein 81-2, HSP81.2, heat shock protein 81.2, HSP90.2, HEAT SHOCK PROTEIN 90.2 |
-0.518 |
0.472 |
| 219 |
ATCG00250 |
ATCG00250
|
TRNE |
-0.514 |
0.537 |
| 220 |
AT1G62390 |
AT1G62390
|
CLMP1, CLUMPED CHLOROPLASTS 1, Phox2, Phox2 |
-0.508 |
-0.082 |
| 221 |
AT3G09440 |
AT3G09440
|
[Heat shock protein 70 (Hsp 70) family protein] |
-0.507 |
0.436 |
| 222 |
AT5G52310 |
AT5G52310
|
COR78, COLD REGULATED 78, LTI140, LTI78, LOW-TEMPERATURE-INDUCED 78, RD29A, RESPONSIVE TO DESSICATION 29A |
-0.500 |
-0.031 |
| 223 |
AT4G29840 |
AT4G29840
|
Pyridoxal-5'-phosphate-dependent enzyme family protein |
-0.496 |
0.049 |
| 224 |
AT5G47110 |
AT5G47110
|
LIL3:2 |
-0.493 |
-0.077 |
| 225 |
AT2G47450 |
AT2G47450
|
CAO, CHAOS, CPSRP43, CHLOROPLAST SIGNAL RECOGNITION PARTICLE 43 |
-0.493 |
0.555 |
| 226 |
AT4G29060 |
AT4G29060
|
emb2726, embryo defective 2726 |
-0.492 |
-0.467 |
| 227 |
AT5G58495 |
AT5G58495
|
[tRNA-Ser (anticodon: CGA)] |
-0.492 |
-0.467 |
| 228 |
ATCG00490 |
ATCG00490
|
RBCL |
-0.487 |
-0.607 |
| 229 |
AT3G54050 |
AT3G54050
|
HCEF1, high cyclic electron flow 1 |
-0.474 |
-0.270 |
| 230 |
AT3G56910 |
AT3G56910
|
PSRP5, plastid-specific 50S ribosomal protein 5 |
-0.465 |
0.009 |
| 231 |
AT5G56010 |
AT5G56010
|
AtHsp90-3, HEAT SHOCK PROTEIN 90-3, AtHsp90.3, HEAT SHOCK PROTEIN 90.3, HSP81-3, heat shock protein 81-3, Hsp81.3, HEAT SHOCK PROTEIN 81.3 |
-0.463 |
0.641 |
| 232 |
AT5G12860 |
AT5G12860
|
DiT1, dicarboxylate transporter 1 |
-0.450 |
-0.293 |
| 233 |
AT1G10522 |
AT1G10522
|
PRIN2, PLASTID REDOX INSENSITIVE 2 |
-0.435 |
1.364 |
| 234 |
AT3G16140 |
AT3G16140
|
PSAH-1, photosystem I subunit H-1 |
-0.428 |
0.465 |
| 235 |
AT4G27440 |
AT4G27440
|
PORB, protochlorophyllide oxidoreductase B |
-0.416 |
1.547 |
| 236 |
AT2G29630 |
AT2G29630
|
PY, PYRIMIDINE REQUIRING, THIC, thiaminC |
-0.411 |
-0.186 |
| 237 |
AT1G61520 |
AT1G61520
|
LHCA3, photosystem I light harvesting complex gene 3 |
-0.403 |
0.380 |
| 238 |
AT2G28000 |
AT2G28000
|
CH-CPN60A, CHLOROPLAST CHAPERONIN 60ALPHA, CPN60A, chaperonin-60alpha, Cpn60alpha1, chaperonin-60alpha1, SLP, SCHLEPPERLESS |
-0.399 |
0.246 |
| 239 |
AT1G15090 |
AT1G15090
|
[tRNA-Asn (anticodon: GTT)] |
-0.398 |
-0.467 |
| 240 |
ATMG00020 |
ATMG00020
|
RRN26, ribosomal RNA26S |
-0.390 |
-0.061 |
| 241 |
AT1G12900 |
AT1G12900
|
GAPA-2, glyceraldehyde 3-phosphate dehydrogenase A subunit 2 |
-0.383 |
-0.029 |
| 242 |
AT1G06680 |
AT1G06680
|
OE23, OXYGEN EVOLVING COMPLEX SUBUNIT 23 KDA, OEE2, OXYGEN-EVOLVING ENHANCER PROTEIN 2, PSBP-1, photosystem II subunit P-1, PSII-P, PHOTOSYSTEM II SUBUNIT P |
-0.378 |
0.620 |
| 243 |
AT4G33010 |
AT4G33010
|
AtGLDP1, glycine decarboxylase P-protein 1, GLDP1, glycine decarboxylase P-protein 1 |
-0.377 |
-0.098 |
| 244 |
AT3G47470 |
AT3G47470
|
CAB4, LHCA4, light-harvesting chlorophyll-protein complex I subunit A4 |
-0.373 |
0.494 |
| 245 |
AT5G23665 |
AT5G23665
|
[tRNA-Gln (anticodon: TTG)] |
-0.365 |
-0.467 |
| 246 |
ATCG00550 |
ATCG00550
|
PSBJ, photosystem II reaction center protein J |
-0.364 |
0.640 |
| 247 |
AT2G37220 |
AT2G37220
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
-0.360 |
-0.097 |
| 248 |
AT5G26000 |
AT5G26000
|
AtTGG1, BGLU38, BETA GLUCOSIDASE 38, TGG1, thioglucoside glucohydrolase 1 |
-0.359 |
-0.122 |
| 249 |
AT3G42240 |
AT3G42240
|
unknown |
-0.356 |
-0.467 |
| 250 |
AT5G01530 |
AT5G01530
|
LHCB4.1, light harvesting complex photosystem II |
-0.355 |
0.588 |
| 251 |
ATCG01060 |
ATCG01060
|
PSAC |
-0.343 |
0.161 |
| 252 |
AT5G38410 |
AT5G38410
|
RBCS3B, Rubisco small subunit 3B |
-0.338 |
-0.028 |
| 253 |
AT4G34950 |
AT4G34950
|
[Major facilitator superfamily protein] |
-0.332 |
0.726 |
| 254 |
AT4G10340 |
AT4G10340
|
LHCB5, light harvesting complex of photosystem II 5 |
-0.324 |
0.446 |