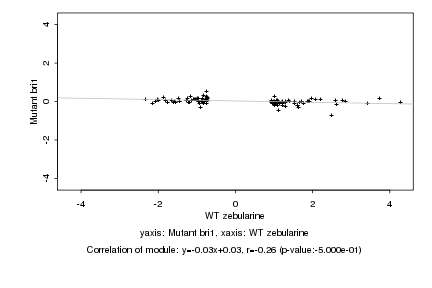
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WT zebularine |
Log2 signal ratio
Mutant bri1 |
| 1 |
AT1G20750 |
AT1G20750
|
[RAD3-like DNA-binding helicase protein] |
4.267 |
-0.018 |
| 2 |
AT1G43800 |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN-DESATURASE6 |
3.728 |
0.180 |
| 3 |
AT4G11050 |
AT4G11050
|
AtGH9C3, glycosyl hydrolase 9C3, GH9C3, glycosyl hydrolase 9C3 |
3.399 |
-0.073 |
| 4 |
AT3G27630 |
AT3G27630
|
SMR7, SIAMESE-RELATED 7 |
2.872 |
no data |
| 5 |
AT2G38600 |
AT2G38600
|
[HAD superfamily, subfamily IIIB acid phosphatase ] |
2.833 |
0.007 |
| 6 |
AT3G49340 |
AT3G49340
|
[Cysteine proteinases superfamily protein] |
2.772 |
0.081 |
| 7 |
AT2G14560 |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
2.609 |
-0.153 |
| 8 |
AT1G12940 |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
2.567 |
0.076 |
| 9 |
AT2G18660 |
AT2G18660
|
AtPNP-A, PNP-A, plant natriuretic peptide A |
2.487 |
-0.687 |
| 10 |
AT2G18193 |
AT2G18193
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
2.486 |
no data |
| 11 |
AT3G01600 |
AT3G01600
|
anac044, NAC domain containing protein 44, NAC044, NAC domain containing protein 44 |
2.445 |
no data |
| 12 |
AT5G15120 |
AT5G15120
|
unknown |
2.185 |
0.127 |
| 13 |
AT3G02550 |
AT3G02550
|
LBD41, LOB domain-containing protein 41 |
2.065 |
0.128 |
| 14 |
AT4G10270 |
AT4G10270
|
[Wound-responsive family protein] |
1.948 |
0.202 |
| 15 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
1.920 |
no data |
| 16 |
AT1G66390 |
AT1G66390
|
ATMYB90, MYB90, myb domain protein 90, PAP2, PRODUCTION OF ANTHOCYANIN PIGMENT 2 |
1.898 |
0.025 |
| 17 |
AT3G57260 |
AT3G57260
|
AtBG2, AtPR2, BG2, BETA-1,3-GLUCANASE 2, BGL2, beta-1,3-glucanase 2, PR-2, PATHOGENESIS-RELATED PROTEIN 2, PR2, PATHOGENESIS-RELATED PROTEIN 2 |
1.879 |
0.023 |
| 18 |
AT5G60250 |
AT5G60250
|
[zinc finger (C3HC4-type RING finger) family protein] |
1.853 |
0.002 |
| 19 |
AT3G46090 |
AT3G46090
|
ZAT7 |
1.804 |
no data |
| 20 |
AT3G22235 |
AT3G22235
|
unknown |
1.791 |
no data |
| 21 |
AT1G65310 |
AT1G65310
|
ATXTH17, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 17, XTH17, xyloglucan endotransglucosylase/hydrolase 17 |
1.740 |
-0.072 |
| 22 |
AT3G58270 |
AT3G58270
|
[Arabidopsis phospholipase-like protein (PEARLI 4) with TRAF-like domain] |
1.729 |
no data |
| 23 |
AT4G22960 |
AT4G22960
|
unknown |
1.686 |
0.050 |
| 24 |
AT5G19100 |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
1.655 |
-0.024 |
| 25 |
AT1G43910 |
AT1G43910
|
[P-loop containing nucleoside triphosphate hydrolases superfamily protein] |
1.624 |
-0.285 |
| 26 |
AT3G47480 |
AT3G47480
|
[Calcium-binding EF-hand family protein] |
1.592 |
-0.173 |
| 27 |
AT5G59240 |
AT5G59240
|
[Ribosomal protein S8e family protein] |
1.584 |
-0.176 |
| 28 |
AT1G24140 |
AT1G24140
|
[Matrixin family protein] |
1.508 |
-0.083 |
| 29 |
AT3G16150 |
AT3G16150
|
ASPGB1, asparaginase B1 |
1.506 |
0.019 |
| 30 |
AT4G39250 |
AT4G39250
|
ATRL1, RAD-like 1, RL1, RAD-like 1, RSM2, RADIALIS-LIKE SANT/MYB 2 |
1.391 |
no data |
| 31 |
AT5G45340 |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
1.381 |
0.053 |
| 32 |
AT2G21650 |
AT2G21650
|
ATRL2, ARABIDOPSIS RAD-LIKE 2, MEE3, MATERNAL EFFECT EMBRYO ARREST 3, RSM1, RADIALIS-LIKE SANT/MYB 1 |
1.373 |
0.073 |
| 33 |
AT3G09922 |
AT3G09922
|
ATIPS1, A. THALIANA INDUCED BY PHOSPHATE STARVATION1, IPS1, INDUCED BY PHOSPHATE STARVATION1 |
1.304 |
no data |
| 34 |
AT3G22740 |
AT3G22740
|
HMT3, homocysteine S-methyltransferase 3 |
1.295 |
0.047 |
| 35 |
AT4G21070 |
AT4G21070
|
ATBRCA1, ARABIDOPSIS THALIANA BREAST CANCER SUSCEPTIBILITY1, BRCA1, breast cancer susceptibility1 |
1.292 |
-0.027 |
| 36 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
1.272 |
-0.247 |
| 37 |
AT2G32680 |
AT2G32680
|
AtRLP23, receptor like protein 23, RLP23, receptor like protein 23 |
1.213 |
-0.023 |
| 38 |
AT1G51380 |
AT1G51380
|
[DEA(D/H)-box RNA helicase family protein] |
1.202 |
-0.159 |
| 39 |
AT2G39330 |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
1.200 |
0.006 |
| 40 |
AT2G18690 |
AT2G18690
|
unknown |
1.197 |
-0.067 |
| 41 |
AT4G25630 |
AT4G25630
|
ATFIB2, FIB2, fibrillarin 2 |
1.197 |
-0.062 |
| 42 |
AT1G21910 |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
1.185 |
-0.022 |
| 43 |
AT1G70260 |
AT1G70260
|
UMAMIT36, Usually multiple acids move in and out Transporters 36 |
1.163 |
no data |
| 44 |
AT3G44750 |
AT3G44750
|
ATHD2A, HD2A, HISTONE DEACETYLASE 2A, HDA3, histone deacetylase 3, HDT1 |
1.138 |
-0.095 |
| 45 |
AT5G10760 |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
1.123 |
-0.097 |
| 46 |
AT5G42800 |
AT5G42800
|
DFR, dihydroflavonol 4-reductase, M318, TT3 |
1.110 |
-0.417 |
| 47 |
AT1G21890 |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
1.092 |
0.036 |
| 48 |
AT3G59670 |
AT3G59670
|
unknown |
1.072 |
-0.171 |
| 49 |
AT3G19680 |
AT3G19680
|
unknown |
1.069 |
0.069 |
| 50 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
1.062 |
-0.044 |
| 51 |
AT2G30670 |
AT2G30670
|
[NAD(P)-binding Rossmann-fold superfamily protein] |
1.056 |
-0.062 |
| 52 |
AT3G07800 |
AT3G07800
|
AtTK1a, TK1a, thymidine kinase 1a |
1.052 |
-0.038 |
| 53 |
AT4G19130 |
AT4G19130
|
[Replication factor-A protein 1-related] |
1.049 |
-0.052 |
| 54 |
AT4G22880 |
AT4G22880
|
ANS, ANTHOCYANIDIN SYNTHASE, LDOX, leucoanthocyanidin dioxygenase, TDS4, TANNIN DEFICIENT SEED 4, TT18 |
1.045 |
no data |
| 55 |
AT5G41080 |
AT5G41080
|
AtGDPD2, GDPD2, glycerophosphodiester phosphodiesterase 2 |
1.045 |
0.094 |
| 56 |
AT3G23370 |
AT3G23370
|
[RNA-binding (RRM/RBD/RNP motifs) family protein] |
1.040 |
no data |
| 57 |
AT2G17040 |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
1.021 |
-0.012 |
| 58 |
AT2G39510 |
AT2G39510
|
UMAMIT14, Usually multiple acids move in and out Transporters 14 |
1.007 |
0.293 |
| 59 |
AT2G32220 |
AT2G32220
|
[Ribosomal L27e protein family] |
1.000 |
-0.142 |
| 60 |
AT5G48720 |
AT5G48720
|
XRI, X-RAY INDUCED TRANSCRIPT, XRI1, X-RAY INDUCED TRANSCRIPT 1 |
0.997 |
0.017 |
| 61 |
AT4G02330 |
AT4G02330
|
AtPME41, ATPMEPCRB, PME41, pectin methylesterase 41 |
0.990 |
-0.191 |
| 62 |
AT5G20850 |
AT5G20850
|
ATRAD51, RAD51 |
0.986 |
0.008 |
| 63 |
AT5G23020 |
AT5G23020
|
IMS2, 2-isopropylmalate synthase 2, MAM-L, METHYLTHIOALKYMALATE SYNTHASE-LIKE, MAM3 |
0.980 |
-0.062 |
| 64 |
AT4G36580 |
AT4G36580
|
[AAA-type ATPase family protein] |
0.946 |
-0.092 |
| 65 |
AT3G19710 |
AT3G19710
|
BCAT4, branched-chain aminotransferase4 |
0.946 |
-0.077 |
| 66 |
AT5G27120 |
AT5G27120
|
[NOP56-like pre RNA processing ribonucleoprotein] |
0.933 |
no data |
| 67 |
AT3G49320 |
AT3G49320
|
[Metal-dependent protein hydrolase] |
0.932 |
0.001 |
| 68 |
AT2G18328 |
AT2G18328
|
ATRL4, RAD-like 4, RL4, RAD-like 4 |
0.928 |
no data |
| 69 |
AT3G30775 |
AT3G30775
|
AT-POX, ATPDH, ATPOX, ARABIDOPSIS THALIANA PROLINE OXIDASE, ERD5, EARLY RESPONSIVE TO DEHYDRATION 5, PDH1, proline dehydrogenase 1, PRO1, PRODH, PROLINE DEHYDROGENASE |
0.926 |
0.009 |
| 70 |
AT1G76650 |
AT1G76650
|
CML38, calmodulin-like 38 |
0.925 |
0.077 |
| 71 |
AT4G28040 |
AT4G28040
|
UMAMIT33, Usually multiple acids move in and out Transporters 33 |
-2.332 |
0.107 |
| 72 |
AT2G26020 |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-2.168 |
-0.091 |
| 73 |
AT3G14280 |
AT3G14280
|
unknown |
-2.091 |
0.019 |
| 74 |
AT5G42900 |
AT5G42900
|
COR27, cold regulated gene 27 |
-2.045 |
0.109 |
| 75 |
AT4G33980 |
AT4G33980
|
unknown |
-2.008 |
0.061 |
| 76 |
AT4G33467 |
AT4G33467
|
unknown |
-1.991 |
no data |
| 77 |
AT2G33830 |
AT2G33830
|
AtDRM2, DRM2, dormancy associated gene 2 |
-1.876 |
0.254 |
| 78 |
AT1G64110 |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-1.830 |
0.058 |
| 79 |
AT2G18050 |
AT2G18050
|
HIS1-3, histone H1-3 |
-1.783 |
-0.048 |
| 80 |
AT3G47340 |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
-1.673 |
0.060 |
| 81 |
AT2G38465 |
AT2G38465
|
unknown |
-1.625 |
-0.028 |
| 82 |
AT2G42530 |
AT2G42530
|
COR15B, cold regulated 15b |
-1.583 |
0.040 |
| 83 |
AT4G25480 |
AT4G25480
|
ATCBF3, CBF3, C-REPEAT BINDING FACTOR 3, DREB1A, dehydration response element B1A |
-1.577 |
-0.028 |
| 84 |
AT1G68050 |
AT1G68050
|
ADO3, FKF1, flavin-binding, kelch repeat, f box 1 |
-1.512 |
no data |
| 85 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-1.498 |
0.196 |
| 86 |
AT3G52310 |
AT3G52310
|
ABCG27, ATP-binding cassette G27 |
-1.466 |
0.029 |
| 87 |
AT2G15020 |
AT2G15020
|
unknown |
-1.451 |
0.044 |
| 88 |
AT4G34550 |
AT4G34550
|
unknown |
-1.394 |
no data |
| 89 |
AT3G24420 |
AT3G24420
|
[alpha/beta-Hydrolases superfamily protein] |
-1.346 |
no data |
| 90 |
AT1G20070 |
AT1G20070
|
unknown |
-1.269 |
0.092 |
| 91 |
AT2G39800 |
AT2G39800
|
ATP5CS, P5CS1, delta1-pyrroline-5-carboxylate synthase 1 |
-1.249 |
no data |
| 92 |
AT4G35770 |
AT4G35770
|
ATSEN1, ARABIDOPSIS THALIANA SENESCENCE 1, DIN1, DARK INDUCIBLE 1, SEN1, SENESCENCE 1, SEN1, SENESCENCE ASSOCIATED GENE 1 |
-1.243 |
0.179 |
| 93 |
AT5G66400 |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-1.230 |
-0.009 |
| 94 |
AT4G08115 |
AT4G08115
|
[gypsy-like retrotransposon family, has a 7.2e-05 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-1.211 |
no data |
| 95 |
AT1G11210 |
AT1G11210
|
unknown |
-1.200 |
-0.017 |
| 96 |
AT4G33550 |
AT4G33550
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.188 |
0.312 |
| 97 |
AT5G54585 |
AT5G54585
|
unknown |
-1.172 |
no data |
| 98 |
AT1G36060 |
AT1G36060
|
[Integrase-type DNA-binding superfamily protein] |
-1.142 |
0.092 |
| 99 |
AT5G24470 |
AT5G24470
|
APRR5, pseudo-response regulator 5, PRR5, pseudo-response regulator 5 |
-1.097 |
0.128 |
| 100 |
AT3G51400 |
AT3G51400
|
unknown |
-1.076 |
0.107 |
| 101 |
AT3G28153 |
AT3G28153
|
[non-LTR retrotransposon family (LINE), has a 1.3e-30 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
-1.067 |
no data |
| 102 |
AT2G21210 |
AT2G21210
|
SAUR6, SMALL AUXIN UPREGULATED RNA 6 |
-1.054 |
0.137 |
| 103 |
AT1G31173 |
AT1G31173
|
MIR167D, microRNA167D |
-1.020 |
no data |
| 104 |
AT2G05540 |
AT2G05540
|
[Glycine-rich protein family] |
-1.018 |
0.109 |
| 105 |
AT5G39610 |
AT5G39610
|
ANAC092, Arabidopsis NAC domain containing protein 92, ATNAC2, NAC domain containing protein 2, ATNAC6, NAC domain containing protein 6, NAC2, NAC domain containing protein 2, NAC6, NAC domain containing protein 6, ORE1, ORESARA 1 |
-1.001 |
0.183 |
| 106 |
AT5G54190 |
AT5G54190
|
PORA, protochlorophyllide oxidoreductase A |
-0.999 |
0.087 |
| 107 |
AT3G28270 |
AT3G28270
|
unknown |
-0.977 |
0.015 |
| 108 |
AT1G77380 |
AT1G77380
|
AAP3, amino acid permease 3, ATAAP3 |
-0.970 |
0.019 |
| 109 |
AT2G33380 |
AT2G33380
|
AtCLO3, Arabidopsis thaliana caleosin 3, CLO-3, caleosin 3, CLO3, caleosin 3, RD20, RESPONSIVE TO DESSICATION 20 |
-0.964 |
0.202 |
| 110 |
AT1G22500 |
AT1G22500
|
AtATL15, Arabidopsis thaliana Arabidopsis toxicos en levadura 15, ATL15, Arabidopsis toxicos en levadura 15 |
-0.959 |
0.062 |
| 111 |
AT1G02820 |
AT1G02820
|
AtLEA3, LEA3, late embryogenesis abundant 3 |
-0.947 |
-0.053 |
| 112 |
AT3G18950 |
AT3G18950
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.937 |
no data |
| 113 |
AT3G09450 |
AT3G09450
|
unknown |
-0.916 |
no data |
| 114 |
AT1G02850 |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-0.907 |
-0.282 |
| 115 |
AT1G76410 |
AT1G76410
|
ATL8 |
-0.897 |
0.087 |
| 116 |
AT5G40720 |
AT5G40720
|
unknown |
-0.887 |
no data |
| 117 |
AT5G56870 |
AT5G56870
|
BGAL4, beta-galactosidase 4 |
-0.882 |
0.013 |
| 118 |
AT1G52342 |
AT1G52342
|
unknown |
-0.878 |
no data |
| 119 |
AT2G22200 |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
-0.875 |
0.004 |
| 120 |
AT1G48330 |
AT1G48330
|
unknown |
-0.854 |
0.179 |
| 121 |
AT2G30420 |
AT2G30420
|
ETC2, ENHANCER OF TRY AND CPC 2 |
-0.840 |
no data |
| 122 |
AT2G42540 |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-0.835 |
-0.095 |
| 123 |
AT5G26200 |
AT5G26200
|
[Mitochondrial substrate carrier family protein] |
-0.832 |
0.362 |
| 124 |
AT4G36850 |
AT4G36850
|
[PQ-loop repeat family protein / transmembrane family protein] |
-0.817 |
no data |
| 125 |
AT1G80570 |
AT1G80570
|
[RNI-like superfamily protein] |
-0.813 |
0.005 |
| 126 |
AT1G73480 |
AT1G73480
|
[alpha/beta-Hydrolases superfamily protein] |
-0.804 |
0.041 |
| 127 |
AT3G20395 |
AT3G20395
|
[RING/U-box superfamily protein] |
-0.798 |
no data |
| 128 |
AT1G48100 |
AT1G48100
|
[Pectin lyase-like superfamily protein] |
-0.796 |
0.149 |
| 129 |
AT3G19030 |
AT3G19030
|
unknown |
-0.792 |
0.108 |
| 130 |
AT3G26165 |
AT3G26165
|
CYP71B18, cytochrome P450, family 71, subfamily B, polypeptide 18 |
-0.791 |
no data |
| 131 |
AT4G16146 |
AT4G16146
|
[cAMP-regulated phosphoprotein 19-related protein] |
-0.775 |
-0.061 |
| 132 |
AT3G62550 |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.773 |
0.297 |
| 133 |
AT4G32340 |
AT4G32340
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-0.770 |
0.134 |
| 134 |
AT4G26530 |
AT4G26530
|
AtFBA5, FBA5, fructose-bisphosphate aldolase 5 |
-0.766 |
0.552 |
| 135 |
AT1G70420 |
AT1G70420
|
unknown |
-0.762 |
0.057 |
| 136 |
AT1G72830 |
AT1G72830
|
ATHAP2C, HAP2C, NF-YA3, nuclear factor Y, subunit A3 |
-0.758 |
0.034 |
| 137 |
AT1G08650 |
AT1G08650
|
ATPPCK1, PHOSPHOENOLPYRUVATE CARBOXYLASE KINASE 1, PPCK1, phosphoenolpyruvate carboxylase kinase 1 |
-0.754 |
0.113 |
| 138 |
AT3G06500 |
AT3G06500
|
A/N-InvC, alkaline/neutral invertase C |
-0.749 |
0.087 |
| 139 |
AT1G62510 |
AT1G62510
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-0.748 |
0.173 |
| 140 |
AT4G19170 |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-0.745 |
0.252 |