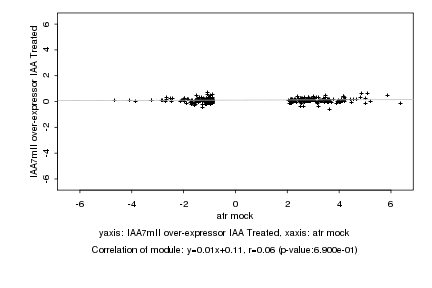
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atr mock |
Log2 signal ratio
IAA7mII over-expressor IAA Treated |
| 1 |
AT3G54580 |
AT3G54580
|
[Proline-rich extensin-like family protein] |
6.353 |
-0.096 |
| 2 |
AT1G30370 |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
5.863 |
0.506 |
| 3 |
AT3G56790 |
AT3G56790
|
[RNA splicing factor-related] |
5.540 |
no data |
| 4 |
AT1G56250 |
AT1G56250
|
AtPP2-B14, phloem protein 2-B14, PP2-B14, phloem protein 2-B14, VBF, VIP1-binding F-box protein |
5.457 |
no data |
| 5 |
AT4G31950 |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
5.173 |
0.023 |
| 6 |
AT3G12910 |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
5.084 |
0.669 |
| 7 |
AT1G07160 |
AT1G07160
|
[Protein phosphatase 2C family protein] |
5.016 |
0.240 |
| 8 |
AT3G54590 |
AT3G54590
|
ATHRGP1, hydroxyproline-rich glycoprotein, HRGP1, hydroxyproline-rich glycoprotein |
4.982 |
-0.080 |
| 9 |
AT3G28550 |
AT3G28550
|
[Proline-rich extensin-like family protein] |
4.951 |
no data |
| 10 |
AT3G29000 |
AT3G29000
|
[Calcium-binding EF-hand family protein] |
4.892 |
no data |
| 11 |
AT1G14540 |
AT1G14540
|
PER4, peroxidase 4 |
4.860 |
0.624 |
| 12 |
AT3G01830 |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
4.787 |
0.332 |
| 13 |
AT3G23230 |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
4.651 |
0.187 |
| 14 |
AT5G11140 |
AT5G11140
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
4.650 |
no data |
| 15 |
AT2G37430 |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
4.527 |
0.176 |
| 16 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
4.456 |
-0.042 |
| 17 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
4.425 |
0.206 |
| 18 |
AT3G48340 |
AT3G48340
|
CEP2, cysteine endopeptidase 2 |
4.390 |
no data |
| 19 |
AT2G32200 |
AT2G32200
|
unknown |
4.231 |
no data |
| 20 |
AT4G30280 |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
4.224 |
0.182 |
| 21 |
AT5G22520 |
AT5G22520
|
unknown |
4.208 |
no data |
| 22 |
AT5G06640 |
AT5G06640
|
EXT10, extensin 10 |
4.207 |
0.321 |
| 23 |
AT1G56240 |
AT1G56240
|
AtPP2-B13, phloem protein 2-B13, PP2-B13, phloem protein 2-B13 |
4.206 |
no data |
| 24 |
AT3G02840 |
AT3G02840
|
[ARM repeat superfamily protein] |
4.201 |
0.040 |
| 25 |
AT1G71520 |
AT1G71520
|
[Integrase-type DNA-binding superfamily protein] |
4.170 |
no data |
| 26 |
AT1G66090 |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
4.154 |
0.460 |
| 27 |
AT3G49130 |
AT3G49130
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
4.148 |
0.411 |
| 28 |
AT5G47850 |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
4.143 |
0.143 |
| 29 |
AT5G64905 |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
4.142 |
0.131 |
| 30 |
AT3G25250 |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
4.122 |
0.058 |
| 31 |
AT5G46890 |
AT5G46890
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
4.085 |
no data |
| 32 |
AT1G22810 |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
4.057 |
0.176 |
| 33 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
4.054 |
0.228 |
| 34 |
AT5G03210 |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
4.045 |
-0.008 |
| 35 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
4.022 |
no data |
| 36 |
AT5G24110 |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
3.987 |
-0.002 |
| 37 |
AT5G65080 |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
3.945 |
0.118 |
| 38 |
AT1G50750 |
AT1G50750
|
[Plant mobile domain protein family] |
3.926 |
0.036 |
| 39 |
AT4G22710 |
AT4G22710
|
CYP706A2, cytochrome P450, family 706, subfamily A, polypeptide 2 |
3.916 |
no data |
| 40 |
AT1G60190 |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
3.894 |
-0.100 |
| 41 |
AT3G46090 |
AT3G46090
|
ZAT7 |
3.845 |
no data |
| 42 |
AT1G42980 |
AT1G42980
|
[Actin-binding FH2 (formin homology 2) family protein] |
3.836 |
no data |
| 43 |
AT1G66270 |
AT1G66270
|
BGLU21 |
3.799 |
no data |
| 44 |
AT4G13390 |
AT4G13390
|
EXT12, extensin 12 |
3.781 |
0.173 |
| 45 |
AT4G14370 |
AT4G14370
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.758 |
no data |
| 46 |
AT1G05675 |
AT1G05675
|
[UDP-Glycosyltransferase superfamily protein] |
3.741 |
no data |
| 47 |
AT2G32030 |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
3.686 |
-0.013 |
| 48 |
AT5G22380 |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
3.643 |
0.074 |
| 49 |
AT1G21120 |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
3.616 |
-0.606 |
| 50 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
3.613 |
0.165 |
| 51 |
AT1G30190 |
AT1G30190
|
unknown |
3.579 |
no data |
| 52 |
AT1G53080 |
AT1G53080
|
[Legume lectin family protein] |
3.568 |
0.168 |
| 53 |
AT1G58420 |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
3.566 |
-0.043 |
| 54 |
AT5G47960 |
AT5G47960
|
ATRABA4C, RAB GTPase homolog A4C, RABA4C, RAB GTPase homolog A4C, SMG1, SMALL MOLECULAR WEIGHT G-PROTEIN 1 |
3.537 |
0.001 |
| 55 |
AT2G41280 |
AT2G41280
|
ATM10, M10 |
3.534 |
0.221 |
| 56 |
AT2G31345 |
AT2G31345
|
unknown |
3.503 |
no data |
| 57 |
AT2G20880 |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
3.487 |
0.066 |
| 58 |
AT1G56060 |
AT1G56060
|
unknown |
3.462 |
0.321 |
| 59 |
AT5G09876 |
AT5G09876
|
unknown |
3.457 |
no data |
| 60 |
AT1G78410 |
AT1G78410
|
[VQ motif-containing protein] |
3.457 |
0.172 |
| 61 |
AT5G45630 |
AT5G45630
|
unknown |
3.453 |
0.524 |
| 62 |
AT1G01310 |
AT1G01310
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
3.442 |
0.010 |
| 63 |
AT2G32140 |
AT2G32140
|
[transmembrane receptors] |
3.423 |
0.204 |
| 64 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
3.421 |
-0.054 |
| 65 |
AT2G32210 |
AT2G32210
|
unknown |
3.415 |
no data |
| 66 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
3.414 |
-0.040 |
| 67 |
AT2G24600 |
AT2G24600
|
[Ankyrin repeat family protein] |
3.404 |
-0.019 |
| 68 |
AT1G33750 |
AT1G33750
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
3.385 |
0.286 |
| 69 |
AT4G14450 |
AT4G14450
|
unknown |
3.358 |
no data |
| 70 |
AT1G21326 |
AT1G21326
|
[VQ motif-containing protein] |
3.334 |
no data |
| 71 |
AT4G18540 |
AT4G18540
|
unknown |
3.332 |
0.100 |
| 72 |
AT1G47405 |
AT1G47405
|
[pseudogene] |
3.320 |
no data |
| 73 |
AT3G54150 |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
3.305 |
0.078 |
| 74 |
AT1G73510 |
AT1G73510
|
unknown |
3.302 |
0.012 |
| 75 |
AT1G43800 |
AT1G43800
|
FTM1, FLORAL TRANSITION AT THE MERISTEM1, SAD6, STEAROYL-ACYL CARRIER PROTEIN-DESATURASE6 |
3.280 |
0.120 |
| 76 |
AT1G20310 |
AT1G20310
|
unknown |
3.275 |
0.033 |
| 77 |
AT1G79680 |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
3.213 |
0.110 |
| 78 |
AT5G52760 |
AT5G52760
|
[Copper transport protein family] |
3.201 |
0.346 |
| 79 |
AT5G28610 |
AT5G28610
|
unknown |
3.193 |
-0.360 |
| 80 |
AT1G29970 |
AT1G29970
|
RPL18AA, 60S ribosomal protein L18A-1 |
3.166 |
0.061 |
| 81 |
AT3G46080 |
AT3G46080
|
[C2H2-type zinc finger family protein] |
3.157 |
no data |
| 82 |
AT4G30170 |
AT4G30170
|
[Peroxidase family protein] |
3.144 |
-0.017 |
| 83 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
3.139 |
-0.103 |
| 84 |
AT1G21110 |
AT1G21110
|
IGMT3, indole glucosinolate O-methyltransferase 3 |
3.127 |
no data |
| 85 |
AT1G05575 |
AT1G05575
|
unknown |
3.112 |
0.333 |
| 86 |
AT5G56960 |
AT5G56960
|
[basic helix-loop-helix (bHLH) DNA-binding family protein] |
3.100 |
no data |
| 87 |
AT5G05300 |
AT5G05300
|
unknown |
3.074 |
0.078 |
| 88 |
AT1G60960 |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
3.046 |
0.257 |
| 89 |
AT4G29780 |
AT4G29780
|
unknown |
3.022 |
0.155 |
| 90 |
AT1G16420 |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
3.020 |
0.135 |
| 91 |
AT1G30390 |
AT1G30390
|
[non-LTR retrotransposon family (LINE), has a 1.9e-17 P-value blast match to GB:AAB41224 ORF2 (LINE-element) (Rattus norvegicus)] |
3.016 |
no data |
| 92 |
AT2G44840 |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
2.992 |
0.154 |
| 93 |
AT3G28340 |
AT3G28340
|
GATL10, galacturonosyltransferase-like 10, GolS8, galactinol synthase 8 |
2.983 |
0.114 |
| 94 |
AT4G11470 |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
2.982 |
0.422 |
| 95 |
AT3G50930 |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
2.980 |
0.275 |
| 96 |
AT1G35210 |
AT1G35210
|
unknown |
2.966 |
0.159 |
| 97 |
AT2G33710 |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
2.951 |
0.131 |
| 98 |
AT1G29680 |
AT1G29680
|
unknown |
2.942 |
0.123 |
| 99 |
AT2G35710 |
AT2G35710
|
PGSIP7, plant glycogenin-like starch initiation protein 7 |
2.940 |
0.276 |
| 100 |
AT2G46400 |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
2.938 |
0.106 |
| 101 |
AT4G15975 |
AT4G15975
|
[RING/U-box superfamily protein] |
2.928 |
no data |
| 102 |
AT3G55840 |
AT3G55840
|
[Hs1pro-1 protein] |
2.927 |
0.188 |
| 103 |
AT5G39670 |
AT5G39670
|
[Calcium-binding EF-hand family protein] |
2.919 |
0.240 |
| 104 |
AT4G21390 |
AT4G21390
|
B120 |
2.883 |
0.125 |
| 105 |
AT2G38470 |
AT2G38470
|
ATWRKY33, WRKY DNA-BINDING PROTEIN 33, WRKY33, WRKY DNA-binding protein 33 |
2.865 |
0.071 |
| 106 |
AT5G27420 |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
2.845 |
0.238 |
| 107 |
AT4G17490 |
AT4G17490
|
ATERF6, ethylene responsive element binding factor 6, ERF-6-6, ERF6, ethylene responsive element binding factor 6 |
2.835 |
0.079 |
| 108 |
AT1G16130 |
AT1G16130
|
WAKL2, wall associated kinase-like 2 |
2.817 |
0.365 |
| 109 |
AT1G73805 |
AT1G73805
|
SARD1, SAR Deficient 1 |
2.816 |
0.217 |
| 110 |
AT3G48650 |
AT3G48650
|
[pseudogene] |
2.794 |
0.074 |
| 111 |
AT3G63060 |
AT3G63060
|
EDL3, EID1-like 3 |
2.790 |
0.201 |
| 112 |
AT4G39580 |
AT4G39580
|
[Galactose oxidase/kelch repeat superfamily protein] |
2.789 |
no data |
| 113 |
AT1G24140 |
AT1G24140
|
[Matrixin family protein] |
2.771 |
0.154 |
| 114 |
AT4G24110 |
AT4G24110
|
unknown |
2.770 |
0.078 |
| 115 |
AT3G10930 |
AT3G10930
|
unknown |
2.770 |
0.093 |
| 116 |
AT1G19020 |
AT1G19020
|
unknown |
2.730 |
0.038 |
| 117 |
AT5G47230 |
AT5G47230
|
ATERF-5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR- 5, ATERF5, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 5, AtMACD1, ERF102, ERF5, ethylene responsive element binding factor 5 |
2.715 |
0.200 |
| 118 |
AT5G52050 |
AT5G52050
|
[MATE efflux family protein] |
2.711 |
0.070 |
| 119 |
AT5G41740 |
AT5G41740
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.707 |
no data |
| 120 |
AT1G10560 |
AT1G10560
|
ATPUB18, ARABIDOPSIS THALIANA PLANT U-BOX 18, PUB18, plant U-box 18 |
2.704 |
0.109 |
| 121 |
AT5G46295 |
AT5G46295
|
unknown |
2.690 |
0.034 |
| 122 |
AT5G13990 |
AT5G13990
|
ATEXO70C2, exocyst subunit exo70 family protein C2, EXO70C2, exocyst subunit exo70 family protein C2 |
2.677 |
0.145 |
| 123 |
AT2G40140 |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
2.672 |
0.084 |
| 124 |
AT1G17380 |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
2.664 |
0.382 |
| 125 |
AT4G40080 |
AT4G40080
|
[ENTH/ANTH/VHS superfamily protein] |
2.658 |
-0.003 |
| 126 |
AT3G55980 |
AT3G55980
|
ATSZF1, SZF1, salt-inducible zinc finger 1 |
2.657 |
-0.035 |
| 127 |
AT3G14440 |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
2.624 |
-0.335 |
| 128 |
AT4G37290 |
AT4G37290
|
unknown |
2.596 |
0.076 |
| 129 |
AT5G66650 |
AT5G66650
|
unknown |
2.570 |
0.239 |
| 130 |
AT1G61340 |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
2.561 |
0.217 |
| 131 |
AT5G66640 |
AT5G66640
|
DAR3, DA1-related protein 3 |
2.554 |
0.198 |
| 132 |
AT5G20230 |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
2.551 |
0.183 |
| 133 |
AT4G39670 |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
2.537 |
0.262 |
| 134 |
AT5G20260 |
AT5G20260
|
[Exostosin family protein] |
2.532 |
no data |
| 135 |
AT1G28370 |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
2.522 |
-0.149 |
| 136 |
AT3G45960 |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
2.519 |
0.104 |
| 137 |
AT3G09870 |
AT3G09870
|
SAUR48, SMALL AUXIN UPREGULATED RNA 48 |
2.498 |
0.042 |
| 138 |
AT5G45340 |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
2.496 |
0.009 |
| 139 |
AT5G48010 |
AT5G48010
|
AtTHAS1, Arabidopsis thaliana thalianol synthase 1, THAS, THALIANOL SYNTHASE, THAS1, thalianol synthase 1 |
2.496 |
0.263 |
| 140 |
AT4G04223 |
AT4G04223
|
other RNA |
2.495 |
no data |
| 141 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
2.487 |
-0.374 |
| 142 |
AT2G24610 |
AT2G24610
|
ATCNGC14, cyclic nucleotide-gated channel 14, CNGC14, cyclic nucleotide-gated channel 14 |
2.487 |
0.286 |
| 143 |
AT3G52400 |
AT3G52400
|
ATSYP122, SYP122, syntaxin of plants 122 |
2.481 |
0.074 |
| 144 |
AT1G07135 |
AT1G07135
|
[glycine-rich protein] |
2.466 |
no data |
| 145 |
AT2G30615 |
AT2G30615
|
unknown |
2.466 |
no data |
| 146 |
AT5G65070 |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
2.458 |
0.018 |
| 147 |
AT5G63130 |
AT5G63130
|
[Octicosapeptide/Phox/Bem1p family protein] |
2.454 |
0.149 |
| 148 |
AT3G45660 |
AT3G45660
|
[Major facilitator superfamily protein] |
2.439 |
no data |
| 149 |
AT1G17147 |
AT1G17147
|
[VQ motif-containing protein] |
2.427 |
no data |
| 150 |
AT5G65600 |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
2.419 |
0.115 |
| 151 |
AT5G51190 |
AT5G51190
|
[Integrase-type DNA-binding superfamily protein] |
2.411 |
0.273 |
| 152 |
AT2G22880 |
AT2G22880
|
[VQ motif-containing protein] |
2.407 |
0.140 |
| 153 |
AT1G26380 |
AT1G26380
|
[FAD-binding Berberine family protein] |
2.361 |
0.449 |
| 154 |
AT5G26290 |
AT5G26290
|
[TRAF-like family protein] |
2.339 |
no data |
| 155 |
AT2G35658 |
AT2G35658
|
unknown |
2.337 |
no data |
| 156 |
AT1G18570 |
AT1G18570
|
AtMYB51, myb domain protein 51, BW51A, BW51B, HIG1, HIGH INDOLIC GLUCOSINOLATE 1, MYB51, myb domain protein 51 |
2.330 |
-0.055 |
| 157 |
AT2G17040 |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
2.301 |
0.170 |
| 158 |
AT2G39650 |
AT2G39650
|
unknown |
2.268 |
0.017 |
| 159 |
AT5G04340 |
AT5G04340
|
C2H2, CZF2, COLD INDUCED ZINC FINGER PROTEIN 2, ZAT6, zinc finger of Arabidopsis thaliana 6 |
2.246 |
0.165 |
| 160 |
AT1G24580 |
AT1G24580
|
[RING/U-box superfamily protein] |
2.235 |
no data |
| 161 |
AT3G11840 |
AT3G11840
|
PUB24, plant U-box 24 |
2.213 |
0.101 |
| 162 |
AT1G17420 |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
2.194 |
0.226 |
| 163 |
AT3G08720 |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
2.185 |
0.111 |
| 164 |
AT5G25250 |
AT5G25250
|
FLOT1, flotillin 1 |
2.183 |
no data |
| 165 |
AT1G63820 |
AT1G63820
|
[CCT motif family protein] |
2.181 |
-0.005 |
| 166 |
AT5G59820 |
AT5G59820
|
AtZAT12, RHL41, RESPONSIVE TO HIGH LIGHT 41, ZAT12 |
2.178 |
0.069 |
| 167 |
AT3G23250 |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
2.148 |
0.206 |
| 168 |
AT5G57010 |
AT5G57010
|
[calmodulin-binding family protein] |
2.146 |
no data |
| 169 |
AT5G47990 |
AT5G47990
|
CYP705A5, cytochrome P450, family 705, subfamily A, polypeptide 5, THAD, THALIAN-DIOL DESATURASE, THAD1, THALIAN-DIOL DESATURASE |
2.145 |
0.210 |
| 170 |
AT1G59590 |
AT1G59590
|
ZCF37 |
2.139 |
-0.110 |
| 171 |
AT1G27770 |
AT1G27770
|
ACA1, autoinhibited Ca2+-ATPase 1, PEA1, PLASTID ENVELOPE ATPASE 1 |
2.119 |
0.041 |
| 172 |
AT5G35735 |
AT5G35735
|
[Auxin-responsive family protein] |
2.109 |
0.145 |
| 173 |
AT2G40095 |
AT2G40095
|
[Alpha/beta hydrolase related protein] |
2.105 |
no data |
| 174 |
AT1G24145 |
AT1G24145
|
unknown |
2.104 |
no data |
| 175 |
AT5G09800 |
AT5G09800
|
[ARM repeat superfamily protein] |
2.102 |
-0.114 |
| 176 |
AT5G39580 |
AT5G39580
|
[Peroxidase superfamily protein] |
2.100 |
0.196 |
| 177 |
AT3G25780 |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
2.071 |
-0.136 |
| 178 |
AT5G54710 |
AT5G54710
|
[Ankyrin repeat family protein] |
2.056 |
no data |
| 179 |
AT4G34150 |
AT4G34150
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
2.040 |
0.150 |
| 180 |
AT1G53625 |
AT1G53625
|
unknown |
2.030 |
no data |
| 181 |
AT1G61470 |
AT1G61470
|
[Polynucleotidyl transferase, ribonuclease H-like superfamily protein] |
2.029 |
no data |
| 182 |
AT4G37990 |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
-4.667 |
0.088 |
| 183 |
AT4G08093 |
AT4G08093
|
[expressed protein, blastp match of 44% identity and 9.5e-07 P-value to GP|14091850|gb|AAK53853.1|AC016781_7|AC016781 Hypothetical protein {Oryza sativa}] |
-4.481 |
no data |
| 184 |
AT1G53480 |
AT1G53480
|
ATMRD1, ARABIDOPSIS MTO 1 RESPONDING DOWN 1, MRD1, MTO 1 RESPONDING DOWN 1 |
-4.328 |
no data |
| 185 |
AT5G44420 |
AT5G44420
|
LCR77, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 77, PDF1.2, plant defensin 1.2, PDF1.2A, PLANT DEFENSIN 1.2A |
-4.111 |
0.139 |
| 186 |
AT2G02990 |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-3.871 |
0.004 |
| 187 |
AT2G26020 |
AT2G26020
|
PDF1.2b, plant defensin 1.2b |
-3.271 |
0.138 |
| 188 |
AT1G10585 |
AT1G10585
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-3.257 |
0.134 |
| 189 |
AT1G62225 |
AT1G62225
|
unknown |
-3.015 |
no data |
| 190 |
AT2G26010 |
AT2G26010
|
PDF1.3, plant defensin 1.3 |
-2.991 |
no data |
| 191 |
AT2G15810 |
AT2G15810
|
[Mutator-like transposase family, has a 1.5e-12 P-value blast match to GB:AAA21566 mudrA of transposon=MuDR (MuDr-element) (Zea mays)] |
-2.876 |
0.127 |
| 192 |
AT5G44430 |
AT5G44430
|
PDF1.2c, plant defensin 1.2C |
-2.869 |
no data |
| 193 |
AT4G12490 |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-2.835 |
0.101 |
| 194 |
AT5G40820 |
AT5G40820
|
ATATR, ARABIDOPSIS THALIANA ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED, ATR, Ataxia telangiectasia-mutated and RAD3-related, ATRAD3 |
-2.718 |
0.009 |
| 195 |
AT3G43715 |
AT3G43715
|
[pseudogene] |
-2.705 |
no data |
| 196 |
AT5G20790 |
AT5G20790
|
unknown |
-2.676 |
0.338 |
| 197 |
AT5G50800 |
AT5G50800
|
AtSWEET13, SWEET13 |
-2.666 |
0.175 |
| 198 |
AT3G20360 |
AT3G20360
|
[TRAF-like family protein] |
-2.650 |
no data |
| 199 |
AT2G11810 |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-2.524 |
0.239 |
| 200 |
AT2G37870 |
AT2G37870
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-2.502 |
0.005 |
| 201 |
AT5G59310 |
AT5G59310
|
LTP4, lipid transfer protein 4 |
-2.445 |
0.237 |
| 202 |
AT1G74290 |
AT1G74290
|
[alpha/beta-Hydrolases superfamily protein] |
-2.156 |
no data |
| 203 |
AT3G55970 |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
-2.138 |
0.010 |
| 204 |
AT1G63580 |
AT1G63580
|
[Receptor-like protein kinase-related family protein] |
-2.086 |
0.094 |
| 205 |
AT4G34550 |
AT4G34550
|
unknown |
-2.040 |
no data |
| 206 |
AT1G79770 |
AT1G79770
|
unknown |
-2.030 |
0.124 |
| 207 |
AT4G17470 |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
-2.009 |
0.203 |
| 208 |
AT5G06510 |
AT5G06510
|
NF-YA10, nuclear factor Y, subunit A10 |
-1.992 |
0.305 |
| 209 |
AT2G36750 |
AT2G36750
|
UGT73C1, UDP-glucosyl transferase 73C1 |
-1.983 |
0.294 |
| 210 |
AT1G21890 |
AT1G21890
|
UMAMIT19, Usually multiple acids move in and out Transporters 19 |
-1.979 |
0.076 |
| 211 |
AT5G20670 |
AT5G20670
|
unknown |
-1.909 |
no data |
| 212 |
AT5G66400 |
AT5G66400
|
ATDI8, ARABIDOPSIS THALIANA DROUGHT-INDUCED 8, RAB18, RESPONSIVE TO ABA 18 |
-1.898 |
-0.091 |
| 213 |
AT3G09450 |
AT3G09450
|
unknown |
-1.878 |
no data |
| 214 |
AT5G48850 |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-1.869 |
0.224 |
| 215 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-1.867 |
0.218 |
| 216 |
AT4G37140 |
AT4G37140
|
ATMES20, ARABIDOPSIS THALIANA METHYL ESTERASE 20, MEE69, MATERNAL EFFECT EMBRYO ARREST 69, MES20, METHYL ESTERASE 20 |
-1.849 |
no data |
| 217 |
AT5G59320 |
AT5G59320
|
LTP3, lipid transfer protein 3 |
-1.835 |
0.163 |
| 218 |
AT5G10760 |
AT5G10760
|
[Eukaryotic aspartyl protease family protein] |
-1.763 |
-0.002 |
| 219 |
AT2G15020 |
AT2G15020
|
unknown |
-1.725 |
0.082 |
| 220 |
AT1G64110 |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
-1.719 |
-0.211 |
| 221 |
AT3G52310 |
AT3G52310
|
ABCG27, ATP-binding cassette G27 |
-1.702 |
0.016 |
| 222 |
AT1G53490 |
AT1G53490
|
HEI10, homolog of human HEI10 ( Enhancer of cell Invasion No.10) |
-1.661 |
-0.036 |
| 223 |
AT5G62360 |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.660 |
0.117 |
| 224 |
AT3G43670 |
AT3G43670
|
[Copper amine oxidase family protein] |
-1.594 |
-0.275 |
| 225 |
AT4G23600 |
AT4G23600
|
CORI3, CORONATINE INDUCED 1, JR2, JASMONIC ACID RESPONSIVE 2 |
-1.587 |
-0.096 |
| 226 |
AT1G31173 |
AT1G31173
|
MIR167D, microRNA167D |
-1.584 |
no data |
| 227 |
AT2G41640 |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-1.575 |
0.138 |
| 228 |
AT1G29395 |
AT1G29395
|
COR413-TM1, COLD REGULATED 314 THYLAKOID MEMBRANE 1, COR413IM1, COLD REGULATED 314 INNER MEMBRANE 1, COR414-TM1, cold regulated 414 thylakoid membrane 1 |
-1.558 |
-0.017 |
| 229 |
AT5G24770 |
AT5G24770
|
ATVSP2, VSP2, vegetative storage protein 2 |
-1.553 |
no data |
| 230 |
AT4G12480 |
AT4G12480
|
EARLI1, EARLY ARABIDOPSIS ALUMINUM INDUCED 1, pEARLI 1 |
-1.545 |
-0.217 |
| 231 |
AT5G26220 |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-1.530 |
0.175 |
| 232 |
AT2G44460 |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-1.525 |
0.483 |
| 233 |
AT2G42530 |
AT2G42530
|
COR15B, cold regulated 15b |
-1.506 |
0.008 |
| 234 |
AT2G37900 |
AT2G37900
|
[Major facilitator superfamily protein] |
-1.476 |
0.236 |
| 235 |
AT3G15310 |
AT3G15310
|
unknown |
-1.476 |
0.071 |
| 236 |
AT5G24780 |
AT5G24780
|
ATVSP1, VSP1, vegetative storage protein 1 |
-1.470 |
no data |
| 237 |
AT3G30122 |
AT3G30122
|
[expressed protein] |
-1.460 |
no data |
| 238 |
AT4G12500 |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.454 |
-0.015 |
| 239 |
AT2G29350 |
AT2G29350
|
SAG13, senescence-associated gene 13 |
-1.453 |
-0.009 |
| 240 |
AT4G15210 |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
-1.439 |
0.060 |
| 241 |
AT5G59780 |
AT5G59780
|
ATMYB59, MYB DOMAIN PROTEIN 59, ATMYB59-1, ATMYB59-2, ATMYB59-3, MYB59, myb domain protein 59 |
-1.437 |
0.093 |
| 242 |
AT1G02850 |
AT1G02850
|
BGLU11, beta glucosidase 11 |
-1.428 |
-0.016 |
| 243 |
AT2G16660 |
AT2G16660
|
[Major facilitator superfamily protein] |
-1.427 |
0.129 |
| 244 |
AT4G37150 |
AT4G37150
|
ATMES9, ARABIDOPSIS THALIANA METHYL ESTERASE 9, MES9, methyl esterase 9 |
-1.420 |
0.367 |
| 245 |
AT4G17220 |
AT4G17220
|
ATMAP70-5, microtubule-associated proteins 70-5, MAP70-5, microtubule-associated proteins 70-5 |
-1.412 |
0.130 |
| 246 |
AT2G44570 |
AT2G44570
|
AtGH9B12, glycosyl hydrolase 9B12, GH9B12, glycosyl hydrolase 9B12 |
-1.402 |
0.053 |
| 247 |
AT5G50950 |
AT5G50950
|
FUM2, FUMARASE 2 |
-1.387 |
no data |
| 248 |
AT1G06160 |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-1.384 |
0.129 |
| 249 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
-1.374 |
no data |
| 250 |
AT2G39330 |
AT2G39330
|
JAL23, jacalin-related lectin 23 |
-1.372 |
0.140 |
| 251 |
AT1G52040 |
AT1G52040
|
ATMBP, MBP1, myrosinase-binding protein 1 |
-1.344 |
no data |
| 252 |
AT5G20150 |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-1.322 |
0.327 |
| 253 |
AT4G34770 |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-1.313 |
0.228 |
| 254 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-1.306 |
0.077 |
| 255 |
AT3G18773 |
AT3G18773
|
[RING/U-box superfamily protein] |
-1.305 |
no data |
| 256 |
AT3G59010 |
AT3G59010
|
PME35, pectin methylesterase 35, PME61, pectin methylesterase 61 |
-1.302 |
0.189 |
| 257 |
AT3G28220 |
AT3G28220
|
[TRAF-like family protein] |
-1.300 |
0.108 |
| 258 |
AT5G11410 |
AT5G11410
|
[Protein kinase superfamily protein] |
-1.292 |
-0.392 |
| 259 |
AT2G32100 |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-1.279 |
-0.155 |
| 260 |
AT1G71140 |
AT1G71140
|
[MATE efflux family protein] |
-1.269 |
0.223 |
| 261 |
AT3G44450 |
AT3G44450
|
unknown |
-1.266 |
0.240 |
| 262 |
AT1G14250 |
AT1G14250
|
[GDA1/CD39 nucleoside phosphatase family protein] |
-1.243 |
no data |
| 263 |
AT1G49200 |
AT1G49200
|
[RING/U-box superfamily protein] |
-1.233 |
0.216 |
| 264 |
AT5G16570 |
AT5G16570
|
GLN1;4, glutamine synthetase 1;4 |
-1.231 |
0.170 |
| 265 |
AT2G04032 |
AT2G04032
|
ZIP7, zinc transporter 7 precursor |
-1.231 |
0.035 |
| 266 |
AT1G13650 |
AT1G13650
|
unknown |
-1.218 |
0.193 |
| 267 |
AT3G45140 |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
-1.216 |
0.110 |
| 268 |
AT1G52342 |
AT1G52342
|
unknown |
-1.196 |
no data |
| 269 |
AT5G54585 |
AT5G54585
|
unknown |
-1.184 |
no data |
| 270 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
-1.180 |
no data |
| 271 |
AT5G61590 |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-1.174 |
0.222 |
| 272 |
AT1G56650 |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
-1.166 |
0.034 |
| 273 |
AT1G19610 |
AT1G19610
|
LCR78, LOW-MOLECULAR-WEIGHT CYSTEINE-RICH 78, PDF1.4 |
-1.165 |
-0.172 |
| 274 |
AT4G15550 |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-1.163 |
-0.010 |
| 275 |
AT5G44575 |
AT5G44575
|
unknown |
-1.155 |
no data |
| 276 |
AT3G53980 |
AT3G53980
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.152 |
-0.217 |
| 277 |
AT2G28780 |
AT2G28780
|
unknown |
-1.149 |
-0.097 |
| 278 |
AT5G52640 |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
-1.140 |
-0.007 |
| 279 |
AT1G29460 |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-1.140 |
0.048 |
| 280 |
AT5G08070 |
AT5G08070
|
TCP17, TCP domain protein 17 |
-1.136 |
0.051 |
| 281 |
AT4G21215 |
AT4G21215
|
unknown |
-1.132 |
no data |
| 282 |
AT2G29500 |
AT2G29500
|
[HSP20-like chaperones superfamily protein] |
-1.128 |
-0.066 |
| 283 |
AT3G59940 |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-1.124 |
-0.076 |
| 284 |
AT5G17300 |
AT5G17300
|
RVE1, REVEILLE 1 |
-1.123 |
0.291 |
| 285 |
AT3G46130 |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-1.120 |
0.115 |
| 286 |
AT5G13320 |
AT5G13320
|
AtGH3.12, GDG1, GH3-LIKE DEFENSE GENE 1, GH3.12, GRETCHEN HAGEN 3.12, PBS3, AVRPPHB SUSCEPTIBLE 3, WIN3, HOPW1-1-INTERACTING 3 |
-1.120 |
0.288 |
| 287 |
AT5G03260 |
AT5G03260
|
LAC11, laccase 11 |
-1.112 |
0.157 |
| 288 |
AT1G54790 |
AT1G54790
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.105 |
-0.009 |
| 289 |
AT5G49740 |
AT5G49740
|
ATFRO7, FERRIC REDUCTION OXIDASE 7, FRO7, ferric reduction oxidase 7 |
-1.103 |
no data |
| 290 |
AT1G29510 |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-1.095 |
0.091 |
| 291 |
AT4G12470 |
AT4G12470
|
AZI1, azelaic acid induced 1 |
-1.093 |
-0.056 |
| 292 |
AT1G11190 |
AT1G11190
|
BFN1, bifunctional nuclease i, ENDO1, ENDONUCLEASE 1 |
-1.087 |
0.576 |
| 293 |
AT4G03400 |
AT4G03400
|
DFL2, DWARF IN LIGHT 2, GH3-10 |
-1.085 |
-0.122 |
| 294 |
AT5G44260 |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-1.082 |
0.092 |
| 295 |
AT3G27200 |
AT3G27200
|
[Cupredoxin superfamily protein] |
-1.080 |
0.109 |
| 296 |
AT2G30750 |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
-1.079 |
0.753 |
| 297 |
AT4G24480 |
AT4G24480
|
[Protein kinase superfamily protein] |
-1.073 |
0.070 |
| 298 |
AT2G40610 |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-1.066 |
0.018 |
| 299 |
AT3G47420 |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-1.066 |
0.204 |
| 300 |
AT4G13800 |
AT4G13800
|
unknown |
-1.065 |
0.153 |
| 301 |
AT1G68570 |
AT1G68570
|
AtNPF3.1, NPF3.1, NRT1/ PTR family 3.1 |
-1.065 |
0.157 |
| 302 |
AT1G12320 |
AT1G12320
|
unknown |
-1.065 |
0.249 |
| 303 |
AT3G45680 |
AT3G45680
|
[Major facilitator superfamily protein] |
-1.063 |
0.100 |
| 304 |
AT3G28740 |
AT3G28740
|
CYP81D11, cytochrome P450, family 81, subfamily D, polypeptide 11 |
-1.061 |
-0.052 |
| 305 |
AT3G17790 |
AT3G17790
|
ATACP5, ATPAP17, PAP17, purple acid phosphatase 17 |
-1.059 |
0.114 |
| 306 |
AT1G23390 |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-1.058 |
-0.029 |
| 307 |
AT2G42540 |
AT2G42540
|
COR15, COR15A, cold-regulated 15a |
-1.057 |
0.161 |
| 308 |
AT3G21670 |
AT3G21670
|
AtNPF6.4, NPF6.4, NRT1/ PTR family 6.4 |
-1.052 |
0.132 |
| 309 |
AT1G71030 |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-1.051 |
0.365 |
| 310 |
AT1G13670 |
AT1G13670
|
unknown |
-1.050 |
0.235 |
| 311 |
AT1G10657 |
AT1G10657
|
unknown |
-1.047 |
no data |
| 312 |
AT1G17745 |
AT1G17745
|
PGDH, 3-phosphoglycerate dehydrogenase, PGDH2, phosphoglycerate dehydrogenase 2 |
-1.044 |
-0.133 |
| 313 |
AT4G19170 |
AT4G19170
|
CCD4, carotenoid cleavage dioxygenase 4, NCED4, nine-cis-epoxycarotenoid dioxygenase 4 |
-1.040 |
0.213 |
| 314 |
AT1G80440 |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-1.038 |
-0.070 |
| 315 |
AT3G28270 |
AT3G28270
|
unknown |
-1.030 |
0.084 |
| 316 |
AT1G66725 |
AT1G66725
|
MIR163, microRNA163 |
-1.026 |
no data |
| 317 |
AT1G68600 |
AT1G68600
|
[Aluminium activated malate transporter family protein] |
-1.026 |
0.158 |
| 318 |
AT1G20160 |
AT1G20160
|
ATSBT5.2 |
-1.024 |
-0.013 |
| 319 |
AT3G59140 |
AT3G59140
|
ABCC10, ATP-binding cassette C10, ATMRP14, multidrug resistance-associated protein 14, MRP14, multidrug resistance-associated protein 14 |
-1.022 |
0.189 |
| 320 |
AT4G30450 |
AT4G30450
|
[glycine-rich protein] |
-1.010 |
0.101 |
| 321 |
AT1G28230 |
AT1G28230
|
ATPUP1, PUP1, purine permease 1 |
-1.010 |
0.336 |
| 322 |
AT5G13170 |
AT5G13170
|
AtSWEET15, SAG29, senescence-associated gene 29, SWEET15 |
-1.002 |
0.213 |
| 323 |
AT1G66540 |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-0.997 |
0.146 |
| 324 |
AT4G12290 |
AT4G12290
|
[Copper amine oxidase family protein] |
-0.993 |
no data |
| 325 |
AT5G45650 |
AT5G45650
|
[subtilase family protein] |
-0.993 |
0.352 |
| 326 |
AT4G33810 |
AT4G33810
|
[Glycosyl hydrolase superfamily protein] |
-0.991 |
0.131 |
| 327 |
AT4G13810 |
AT4G13810
|
AtRLP47, receptor like protein 47, RLP47, receptor like protein 47 |
-0.989 |
0.135 |
| 328 |
AT4G14400 |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.988 |
0.063 |
| 329 |
AT1G74810 |
AT1G74810
|
BOR5, REQUIRES HIGH BORON 5 |
-0.986 |
-0.050 |
| 330 |
AT3G19620 |
AT3G19620
|
[Glycosyl hydrolase family protein] |
-0.982 |
0.074 |
| 331 |
AT5G50160 |
AT5G50160
|
ATFRO8, FERRIC REDUCTION OXIDASE 8, FRO8, ferric reduction oxidase 8 |
-0.980 |
0.048 |
| 332 |
AT5G56550 |
AT5G56550
|
ATOXS3, OXIDATIVE STRESS 3, OXS3, OXIDATIVE STRESS 3 |
-0.979 |
no data |
| 333 |
AT2G38940 |
AT2G38940
|
ATPT2, ARABIDOPSIS THALIANA PHOSPHATE TRANSPORTER 2, PHT1;4, phosphate transporter 1;4 |
-0.979 |
no data |
| 334 |
AT5G02190 |
AT5G02190
|
ATASP38, ARABIDOPSIS THALIANA ASPARTIC PROTEASE 38, EMB24, EMBRYO DEFECTIVE 24, PCS1, PROMOTION OF CELL SURVIVAL 1 |
-0.977 |
0.298 |
| 335 |
AT2G31560 |
AT2G31560
|
unknown |
-0.975 |
no data |
| 336 |
AT1G75490 |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
-0.974 |
-0.221 |
| 337 |
AT2G29940 |
AT2G29940
|
ABCG31, ATP-binding cassette G31, ATPDR3, PLEIOTROPIC DRUG RESISTANCE 3, PDR3, pleiotropic drug resistance 3 |
-0.973 |
0.505 |
| 338 |
AT5G59130 |
AT5G59130
|
[Subtilase family protein] |
-0.950 |
0.089 |
| 339 |
AT2G30230 |
AT2G30230
|
unknown |
-0.945 |
no data |
| 340 |
AT1G52030 |
AT1G52030
|
F-ATMBP, MBP1.2, MBP2, myrosinase-binding protein 2 |
-0.945 |
no data |
| 341 |
AT5G18030 |
AT5G18030
|
[SAUR-like auxin-responsive protein family ] |
-0.943 |
no data |
| 342 |
AT4G17030 |
AT4G17030
|
AT-EXPR, ATEXLB1, expansin-like B1, ATEXPR1, ATHEXP BETA 3.1, EXLB1, expansin-like B1, EXPR |
-0.941 |
0.195 |
| 343 |
AT1G62540 |
AT1G62540
|
FMO GS-OX2, flavin-monooxygenase glucosinolate S-oxygenase 2 |
-0.923 |
0.106 |
| 344 |
AT3G05030 |
AT3G05030
|
ATNHX2, NHX2, sodium hydrogen exchanger 2 |
-0.922 |
0.133 |
| 345 |
AT2G40460 |
AT2G40460
|
[Major facilitator superfamily protein] |
-0.917 |
0.211 |
| 346 |
AT2G18050 |
AT2G18050
|
HIS1-3, histone H1-3 |
-0.917 |
-0.080 |
| 347 |
AT5G37600 |
AT5G37600
|
ATGLN1;1, ARABIDOPSIS GLUTAMINE SYNTHASE 1;1, ATGSR1, ARABIDOPSIS THALIANA GLUTAMINE SYNTHASE CLONE R1, GLN1;1, GLUTAMINE SYNTHASE 1;1, GSR 1, glutamine synthase clone R1 |
-0.916 |
0.185 |
| 348 |
AT3G10525 |
AT3G10525
|
LGO, LOSS OF GIANT CELLS FROM ORGANS, SMR1, SIAMESE RELATED 1 |
-0.913 |
-0.002 |
| 349 |
AT1G66100 |
AT1G66100
|
[Plant thionin] |
-0.909 |
0.075 |
| 350 |
AT2G37760 |
AT2G37760
|
AKR4C8, Aldo-keto reductase family 4 member C8 |
-0.909 |
-0.007 |
| 351 |
AT1G25530 |
AT1G25530
|
[Transmembrane amino acid transporter family protein] |
-0.909 |
0.030 |
| 352 |
AT1G05680 |
AT1G05680
|
UGT74E2, Uridine diphosphate glycosyltransferase 74E2 |
-0.906 |
0.349 |
| 353 |
AT5G18130 |
AT5G18130
|
unknown |
-0.904 |
0.257 |
| 354 |
AT2G29090 |
AT2G29090
|
CYP707A2, cytochrome P450, family 707, subfamily A, polypeptide 2 |
-0.904 |
0.020 |
| 355 |
AT1G13080 |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.904 |
0.615 |
| 356 |
AT5G50335 |
AT5G50335
|
unknown |
-0.901 |
0.110 |
| 357 |
AT3G62270 |
AT3G62270
|
BOR2, REQUIRES HIGH BORON 2 |
-0.900 |
-0.094 |
| 358 |
AT1G20620 |
AT1G20620
|
ATCAT3, CAT3, catalase 3, SEN2, SENESCENCE 2 |
-0.899 |
-0.072 |
| 359 |
AT1G62180 |
AT1G62180
|
5'adenylylphosphosulfate reductase 2 |
-0.898 |
0.003 |
| 360 |
AT1G14430 |
AT1G14430
|
[glyoxal oxidase-related protein] |
-0.893 |
no data |
| 361 |
AT3G11110 |
AT3G11110
|
[RING/U-box superfamily protein] |
-0.892 |
0.137 |