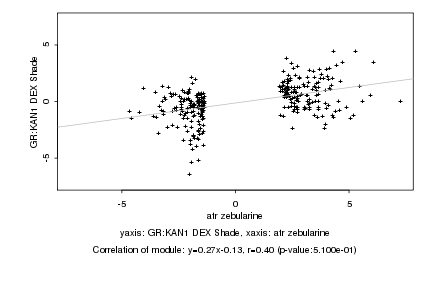
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
atr zebularine |
Log2 signal ratio
GR:KAN1 DEX Shade |
| 1 |
AT4G08091 |
AT4G08091
|
[CACTA-like transposase family (Ptta/En/Spm), has a 4.4e-31 P-value blast match to At5g29026.1/8-244 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
7.249 |
0.061 |
| 2 |
AT3G12910 |
AT3G12910
|
[NAC (No Apical Meristem) domain transcriptional regulator superfamily protein] |
6.035 |
3.516 |
| 3 |
AT3G56790 |
AT3G56790
|
[RNA splicing factor-related] |
5.913 |
0.593 |
| 4 |
AT5G67310 |
AT5G67310
|
CYP81G1, cytochrome P450, family 81, subfamily G, polypeptide 1 |
5.574 |
0.061 |
| 5 |
AT1G60190 |
AT1G60190
|
AtPUB19, PUB19, plant U-box 19 |
5.434 |
1.400 |
| 6 |
AT4G31950 |
AT4G31950
|
CYP82C3, cytochrome P450, family 82, subfamily C, polypeptide 3 |
5.285 |
4.503 |
| 7 |
AT5G03210 |
AT5G03210
|
AtDIP2, DIP2, DBP-interacting protein 2 |
5.163 |
-1.195 |
| 8 |
AT2G20880 |
AT2G20880
|
AtERF53, ERF53, ERF domain 53 |
5.054 |
-1.446 |
| 9 |
AT1G73510 |
AT1G73510
|
unknown |
4.872 |
-0.468 |
| 10 |
AT4G18540 |
AT4G18540
|
unknown |
4.686 |
3.508 |
| 11 |
AT4G22710 |
AT4G22710
|
CYP706A2, cytochrome P450, family 706, subfamily A, polypeptide 2 |
4.618 |
1.799 |
| 12 |
AT1G14540 |
AT1G14540
|
PER4, peroxidase 4 |
4.576 |
-0.778 |
| 13 |
AT1G30370 |
AT1G30370
|
DLAH, DAD1-like acylhydrolase |
4.512 |
0.007 |
| 14 |
AT3G23230 |
AT3G23230
|
AtERF98, AtTDR1, ERF98, Ethylene Response Factor 98, TDR1, Transcriptional Regulator of Defense Response 1 |
4.435 |
3.190 |
| 15 |
AT4G14368 |
AT4G14368
|
[Regulator of chromosome condensation (RCC1) family protein] |
4.367 |
-0.801 |
| 16 |
AT1G55440 |
AT1G55440
|
[Cysteine/Histidine-rich C1 domain family protein] |
4.316 |
-1.405 |
| 17 |
AT1G56060 |
AT1G56060
|
unknown |
4.288 |
4.487 |
| 18 |
AT1G79680 |
AT1G79680
|
ATWAKL10, WAKL10, WALL ASSOCIATED KINASE (WAK)-LIKE 10 |
4.269 |
2.101 |
| 19 |
AT4G25090 |
AT4G25090
|
[Riboflavin synthase-like superfamily protein] |
4.229 |
-1.201 |
| 20 |
AT1G71520 |
AT1G71520
|
[Integrase-type DNA-binding superfamily protein] |
4.202 |
1.449 |
| 21 |
AT1G56240 |
AT1G56240
|
AtPP2-B13, phloem protein 2-B13, PP2-B13, phloem protein 2-B13 |
4.147 |
1.096 |
| 22 |
AT5G24110 |
AT5G24110
|
ATWRKY30, ARABIDOPSIS THALIANA WRKY DNA-BINDING PROTEIN 30, WRKY30, WRKY DNA-binding protein 30 |
4.125 |
2.970 |
| 23 |
AT5G42380 |
AT5G42380
|
CML37, calmodulin like 37 |
4.106 |
1.176 |
| 24 |
AT5G65080 |
AT5G65080
|
AGL68, AGAMOUS-like 68, MAF5, MADS AFFECTING FLOWERING 5 |
4.081 |
-0.343 |
| 25 |
AT1G56250 |
AT1G56250
|
AtPP2-B14, phloem protein 2-B14, PP2-B14, phloem protein 2-B14, VBF, VIP1-binding F-box protein |
4.077 |
1.987 |
| 26 |
AT1G07160 |
AT1G07160
|
[Protein phosphatase 2C family protein] |
4.024 |
2.260 |
| 27 |
AT1G60960 |
AT1G60960
|
ATIRT3, IRON REGULATED TRANSPORTER 3, IRT3, iron regulated transporter 3 |
3.990 |
-0.578 |
| 28 |
AT5G11140 |
AT5G11140
|
[Arabidopsis phospholipase-like protein (PEARLI 4) family] |
3.978 |
2.831 |
| 29 |
AT3G14440 |
AT3G14440
|
ATNCED3, NCED3, nine-cis-epoxycarotenoid dioxygenase 3, SIS7, SUGAR INSENSITIVE 7, STO1, SALT TOLERANT 1 |
3.935 |
-2.002 |
| 30 |
AT1G42980 |
AT1G42980
|
[Actin-binding FH2 (formin homology 2) family protein] |
3.934 |
-0.148 |
| 31 |
AT3G59030 |
AT3G59030
|
ATTT12, A. THALIANA TRANSPARENT TESTA, TT12, TRANSPARENT TESTA 12 |
3.925 |
0.981 |
| 32 |
AT1G22810 |
AT1G22810
|
[Integrase-type DNA-binding superfamily protein] |
3.890 |
-2.298 |
| 33 |
AT3G02840 |
AT3G02840
|
[ARM repeat superfamily protein] |
3.873 |
2.062 |
| 34 |
AT4G21390 |
AT4G21390
|
B120 |
3.789 |
-1.284 |
| 35 |
AT1G05675 |
AT1G05675
|
[UDP-Glycosyltransferase superfamily protein] |
3.750 |
2.450 |
| 36 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
3.743 |
2.040 |
| 37 |
AT3G29000 |
AT3G29000
|
[Calcium-binding EF-hand family protein] |
3.674 |
2.889 |
| 38 |
AT1G66090 |
AT1G66090
|
[Disease resistance protein (TIR-NBS class)] |
3.656 |
0.642 |
| 39 |
AT5G28610 |
AT5G28610
|
unknown |
3.650 |
-0.647 |
| 40 |
AT4G24170 |
AT4G24170
|
[ATP binding microtubule motor family protein] |
3.647 |
-0.469 |
| 41 |
AT1G15990 |
AT1G15990
|
ATCNGC7, cyclic nucleotide gated channel 7, CNGC7, cyclic nucleotide gated channel 7 |
3.647 |
0.061 |
| 42 |
AT4G30280 |
AT4G30280
|
ATXTH18, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 18, XTH18, xyloglucan endotransglucosylase/hydrolase 18 |
3.644 |
1.704 |
| 43 |
AT5G56960 |
AT5G56960
|
[basic helix-loop-helix (bHLH) DNA-binding family protein] |
3.629 |
1.283 |
| 44 |
AT3G49130 |
AT3G49130
|
[SWAP (Suppressor-of-White-APricot)/surp RNA-binding domain-containing protein] |
3.579 |
-1.405 |
| 45 |
AT3G25250 |
AT3G25250
|
AGC2, AGC2-1, AGC2 kinase 1, AtOXI1, OXI1, oxidative signal-inducible1 |
3.565 |
0.581 |
| 46 |
AT1G50750 |
AT1G50750
|
[Plant mobile domain protein family] |
3.553 |
1.657 |
| 47 |
AT1G21110 |
AT1G21110
|
IGMT3, indole glucosinolate O-methyltransferase 3 |
3.550 |
1.000 |
| 48 |
AT3G54150 |
AT3G54150
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
3.499 |
1.241 |
| 49 |
AT5G15660 |
AT5G15660
|
[F-box and associated interaction domains-containing protein] |
3.485 |
0.061 |
| 50 |
AT1G26380 |
AT1G26380
|
[FAD-binding Berberine family protein] |
3.458 |
1.368 |
| 51 |
AT4G28140 |
AT4G28140
|
[Integrase-type DNA-binding superfamily protein] |
3.449 |
-1.157 |
| 52 |
AT1G21120 |
AT1G21120
|
IGMT2, indole glucosinolate O-methyltransferase 2 |
3.444 |
2.196 |
| 53 |
AT1G20310 |
AT1G20310
|
unknown |
3.421 |
2.691 |
| 54 |
AT1G29970 |
AT1G29970
|
RPL18AA, 60S ribosomal protein L18A-1 |
3.383 |
-0.061 |
| 55 |
AT5G59220 |
AT5G59220
|
HAI1, highly ABA-induced PP2C gene 1, SAG113, senescence associated gene 113 |
3.361 |
1.138 |
| 56 |
AT2G37430 |
AT2G37430
|
ZAT11, zinc finger of Arabidopsis thaliana 11 |
3.256 |
-0.692 |
| 57 |
AT5G64905 |
AT5G64905
|
PROPEP3, elicitor peptide 3 precursor |
3.244 |
0.182 |
| 58 |
AT1G56600 |
AT1G56600
|
AtGolS2, galactinol synthase 2, GolS2, galactinol synthase 2 |
3.238 |
-0.146 |
| 59 |
AT1G43000 |
AT1G43000
|
[PLATZ transcription factor family protein] |
3.235 |
2.748 |
| 60 |
AT4G25490 |
AT4G25490
|
ATCBF1, CBF1, C-repeat/DRE binding factor 1, DREB1B, DRE BINDING PROTEIN 1B |
3.223 |
-0.469 |
| 61 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
3.183 |
1.712 |
| 62 |
AT5G64401 |
AT5G64401
|
unknown |
3.179 |
-0.240 |
| 63 |
AT3G01830 |
AT3G01830
|
[Calcium-binding EF-hand family protein] |
3.158 |
2.177 |
| 64 |
AT4G14370 |
AT4G14370
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
3.139 |
1.416 |
| 65 |
AT2G01422 |
AT2G01422
|
|
3.136 |
0.061 |
| 66 |
AT5G65600 |
AT5G65600
|
LecRK-IX.2, L-type lectin receptor kinase IX.2 |
3.132 |
0.561 |
| 67 |
AT3G48630 |
AT3G48630
|
unknown |
3.113 |
0.592 |
| 68 |
AT5G45630 |
AT5G45630
|
unknown |
3.057 |
-0.537 |
| 69 |
AT2G44840 |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
3.033 |
1.408 |
| 70 |
AT4G05170 |
AT4G05170
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
3.023 |
-0.688 |
| 71 |
AT3G48510 |
AT3G48510
|
unknown |
3.004 |
-0.469 |
| 72 |
AT4G14450 |
AT4G14450
|
unknown |
2.977 |
1.535 |
| 73 |
AT1G09930 |
AT1G09930
|
ATOPT2, oligopeptide transporter 2, OPT2, oligopeptide transporter 2 |
2.897 |
0.629 |
| 74 |
AT2G24600 |
AT2G24600
|
[Ankyrin repeat family protein] |
2.865 |
0.672 |
| 75 |
AT2G29100 |
AT2G29100
|
ATGLR2.9, glutamate receptor 2.9, GLR2.9, GLUTAMATE RECEPTOR 2.9, GLR2.9, glutamate receptor 2.9 |
2.840 |
0.612 |
| 76 |
AT1G21326 |
AT1G21326
|
[VQ motif-containing protein] |
2.802 |
2.048 |
| 77 |
AT5G22520 |
AT5G22520
|
unknown |
2.790 |
1.243 |
| 78 |
AT5G41750 |
AT5G41750
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.789 |
2.160 |
| 79 |
AT5G22380 |
AT5G22380
|
anac090, NAC domain containing protein 90, NAC090, NAC domain containing protein 90 |
2.774 |
0.061 |
| 80 |
AT1G78410 |
AT1G78410
|
[VQ motif-containing protein] |
2.725 |
3.178 |
| 81 |
AT3G63060 |
AT3G63060
|
EDL3, EID1-like 3 |
2.724 |
0.279 |
| 82 |
AT1G07430 |
AT1G07430
|
AIP1, AKT1 interacting protein phosphatase 1, AtAIP1, HAI2, highly ABA-induced PP2C gene 2, HON, HONSU (Korean for abnormal drowsiness) |
2.722 |
1.323 |
| 83 |
AT3G45660 |
AT3G45660
|
[Major facilitator superfamily protein] |
2.722 |
-0.636 |
| 84 |
AT5G40800 |
AT5G40800
|
unknown |
2.715 |
-0.468 |
| 85 |
AT5G57220 |
AT5G57220
|
CYP81F2, cytochrome P450, family 81, subfamily F, polypeptide 2 |
2.689 |
-0.947 |
| 86 |
AT2G47140 |
AT2G47140
|
AtSDR5, SDR5, short-chain dehydrogenase reductase 5 |
2.689 |
0.449 |
| 87 |
AT1G16420 |
AT1G16420
|
ATMC8, ARABIDOPSIS THALIANA METACASPASE 8, AtMCP2e, metacaspase 2e, MC8, metacaspase 8, MCP2e, metacaspase 2e |
2.687 |
0.882 |
| 88 |
AT3G56275 |
AT3G56275
|
[expressed protein] |
2.681 |
-0.509 |
| 89 |
AT2G32200 |
AT2G32200
|
unknown |
2.662 |
0.128 |
| 90 |
AT1G73805 |
AT1G73805
|
SARD1, SAR Deficient 1 |
2.639 |
2.346 |
| 91 |
AT2G35658 |
AT2G35658
|
unknown |
2.610 |
0.852 |
| 92 |
AT4G37290 |
AT4G37290
|
unknown |
2.583 |
0.934 |
| 93 |
AT4G39580 |
AT4G39580
|
[Galactose oxidase/kelch repeat superfamily protein] |
2.583 |
0.374 |
| 94 |
AT1G58420 |
AT1G58420
|
[Uncharacterised conserved protein UCP031279] |
2.581 |
1.143 |
| 95 |
AT5G65070 |
AT5G65070
|
AGL69, AGAMOUS-like 69, FCL4, MAF4, MADS AFFECTING FLOWERING 4 |
2.577 |
-0.163 |
| 96 |
AT5G37490 |
AT5G37490
|
[ARM repeat superfamily protein] |
2.575 |
-0.398 |
| 97 |
AT2G32030 |
AT2G32030
|
[Acyl-CoA N-acyltransferases (NAT) superfamily protein] |
2.570 |
1.271 |
| 98 |
AT1G69260 |
AT1G69260
|
AFP1, ABI five binding protein |
2.566 |
-0.643 |
| 99 |
AT1G05575 |
AT1G05575
|
unknown |
2.549 |
-0.797 |
| 100 |
AT1G19020 |
AT1G19020
|
unknown |
2.522 |
0.861 |
| 101 |
AT1G01560 |
AT1G01560
|
ATMPK11, MAP kinase 11, MPK11, MAP kinase 11 |
2.520 |
2.924 |
| 102 |
AT3G11840 |
AT3G11840
|
PUB24, plant U-box 24 |
2.511 |
0.333 |
| 103 |
AT2G33710 |
AT2G33710
|
[Integrase-type DNA-binding superfamily protein] |
2.479 |
0.353 |
| 104 |
AT1G49450 |
AT1G49450
|
[Transducin/WD40 repeat-like superfamily protein] |
2.476 |
-2.356 |
| 105 |
AT5G47850 |
AT5G47850
|
CCR4, CRINKLY4 related 4 |
2.458 |
3.428 |
| 106 |
AT4G15975 |
AT4G15975
|
[RING/U-box superfamily protein] |
2.445 |
0.868 |
| 107 |
AT3G55840 |
AT3G55840
|
[Hs1pro-1 protein] |
2.443 |
1.309 |
| 108 |
AT3G27250 |
AT3G27250
|
unknown |
2.441 |
-0.405 |
| 109 |
AT1G22440 |
AT1G22440
|
[Zinc-binding alcohol dehydrogenase family protein] |
2.438 |
-0.006 |
| 110 |
AT5G27420 |
AT5G27420
|
ATL31, Arabidopsis toxicos en levadura 31, CNI1, carbon/nitrogen insensitive 1 |
2.409 |
1.922 |
| 111 |
AT1G17147 |
AT1G17147
|
[VQ motif-containing protein] |
2.399 |
0.179 |
| 112 |
AT3G48650 |
AT3G48650
|
[pseudogene] |
2.387 |
2.083 |
| 113 |
AT1G73810 |
AT1G73810
|
[Core-2/I-branching beta-1,6-N-acetylglucosaminyltransferase family protein] |
2.374 |
0.513 |
| 114 |
AT4G37370 |
AT4G37370
|
CYP81D8, cytochrome P450, family 81, subfamily D, polypeptide 8 |
2.328 |
2.346 |
| 115 |
AT5G38310 |
AT5G38310
|
unknown |
2.322 |
-0.504 |
| 116 |
AT4G39670 |
AT4G39670
|
[Glycolipid transfer protein (GLTP) family protein] |
2.316 |
1.813 |
| 117 |
AT1G61340 |
AT1G61340
|
AtFBS1, FBS1, F-box stress induced 1 |
2.312 |
0.625 |
| 118 |
AT1G80840 |
AT1G80840
|
ATWRKY40, WRKY40, WRKY DNA-binding protein 40 |
2.303 |
1.082 |
| 119 |
AT2G32210 |
AT2G32210
|
unknown |
2.290 |
1.635 |
| 120 |
AT4G05100 |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
2.288 |
0.534 |
| 121 |
AT5G41550 |
AT5G41550
|
[Disease resistance protein (TIR-NBS-LRR class) family] |
2.282 |
0.637 |
| 122 |
AT3G15518 |
AT3G15518
|
unknown |
2.277 |
1.174 |
| 123 |
AT5G66650 |
AT5G66650
|
unknown |
2.264 |
1.033 |
| 124 |
AT3G50930 |
AT3G50930
|
BCS1, cytochrome BC1 synthesis |
2.256 |
1.301 |
| 125 |
AT4G24570 |
AT4G24570
|
DIC2, dicarboxylate carrier 2 |
2.256 |
1.994 |
| 126 |
AT1G16130 |
AT1G16130
|
WAKL2, wall associated kinase-like 2 |
2.252 |
1.337 |
| 127 |
AT1G64380 |
AT1G64380
|
[Integrase-type DNA-binding superfamily protein] |
2.238 |
0.405 |
| 128 |
AT4G29780 |
AT4G29780
|
unknown |
2.224 |
0.865 |
| 129 |
AT1G28370 |
AT1G28370
|
ATERF11, ERF DOMAIN PROTEIN 11, ERF11, ERF domain protein 11 |
2.214 |
3.877 |
| 130 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
2.197 |
0.576 |
| 131 |
AT5G01100 |
AT5G01100
|
FRB1, FRIABLE 1 |
2.193 |
0.835 |
| 132 |
AT4G01010 |
AT4G01010
|
ATCNGC13, CYCLIC NUCLEOTIDE-GATED CHANNEL 13, CNGC13, cyclic nucleotide-gated channel 13 |
2.193 |
0.459 |
| 133 |
AT4G11470 |
AT4G11470
|
CRK31, cysteine-rich RLK (RECEPTOR-like protein kinase) 31 |
2.187 |
1.154 |
| 134 |
AT3G08720 |
AT3G08720
|
ATPK19, Arabidopsis thaliana protein kinase 19, ATPK2, ARABIDOPSIS THALIANA PROTEIN KINASE 2, ATS6K2, ARABIDOPSIS THALIANA SERINE/THREONINE PROTEIN KINASE 2, S6K2, serine/threonine protein kinase 2 |
2.143 |
-0.498 |
| 135 |
AT2G40140 |
AT2G40140
|
ATSZF2, CZF1, SZF2, (SALT-INDUCIBLE ZINC FINGER 2, ZFAR1 |
2.134 |
0.540 |
| 136 |
AT2G46400 |
AT2G46400
|
ATWRKY46, WRKY DNA-BINDING PROTEIN 46, WRKY46, WRKY DNA-binding protein 46 |
2.130 |
1.802 |
| 137 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
2.121 |
1.077 |
| 138 |
AT1G64110 |
AT1G64110
|
DAA1, DUO1-activated ATPase 1 |
2.118 |
0.358 |
| 139 |
AT1G02400 |
AT1G02400
|
ATGA2OX4, Arabidopsis thaliana gibberellin 2-oxidase 4, ATGA2OX6, ARABIDOPSIS THALIANA GIBBERELLIN 2-OXIDASE 6, DTA1, DOWNSTREAM TARGET OF AGL15 1, GA2OX6, gibberellin 2-oxidase 6 |
2.112 |
-1.291 |
| 140 |
AT5G05300 |
AT5G05300
|
unknown |
2.077 |
1.472 |
| 141 |
AT3G45960 |
AT3G45960
|
ATEXLA3, expansin-like A3, ATEXPL3, ATHEXP BETA 2.3, EXLA3, expansin-like A3, EXPL3 |
2.075 |
2.702 |
| 142 |
AT2G32140 |
AT2G32140
|
[transmembrane receptors] |
2.070 |
1.135 |
| 143 |
AT1G07135 |
AT1G07135
|
[glycine-rich protein] |
2.056 |
1.373 |
| 144 |
AT4G18195 |
AT4G18195
|
ATPUP8, ARABIDOPSIS THALIANA PURINE PERMEASE 8, PUP8, purine permease 8 |
2.050 |
1.692 |
| 145 |
AT2G22880 |
AT2G22880
|
[VQ motif-containing protein] |
2.040 |
0.946 |
| 146 |
AT5G20230 |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
1.969 |
1.252 |
| 147 |
AT1G30700 |
AT1G30700
|
[FAD-binding Berberine family protein] |
1.965 |
-1.164 |
| 148 |
AT5G25930 |
AT5G25930
|
[Protein kinase family protein with leucine-rich repeat domain] |
1.945 |
0.953 |
| 149 |
AT3G10930 |
AT3G10930
|
unknown |
1.936 |
1.390 |
| 150 |
AT1G23110 |
AT1G23110
|
unknown |
-4.698 |
-0.812 |
| 151 |
AT5G20790 |
AT5G20790
|
unknown |
-4.616 |
-1.464 |
| 152 |
AT3G09922 |
AT3G09922
|
ATIPS1, A. THALIANA INDUCED BY PHOSPHATE STARVATION1, IPS1, INDUCED BY PHOSPHATE STARVATION1 |
-4.263 |
-0.948 |
| 153 |
AT4G08093 |
AT4G08093
|
[expressed protein, blastp match of 44% identity and 9.5e-07 P-value to GP|14091850|gb|AAK53853.1|AC016781_7|AC016781 Hypothetical protein {Oryza sativa}] |
-4.055 |
1.218 |
| 154 |
AT2G11810 |
AT2G11810
|
ATMGD3, MGD3, MONOGALACTOSYL DIACYLGLYCEROL SYNTHASE 3, MGDC, monogalactosyldiacylglycerol synthase type C |
-3.628 |
-1.298 |
| 155 |
AT5G48850 |
AT5G48850
|
ATSDI1, SULPHUR DEFICIENCY-INDUCED 1 |
-3.531 |
0.820 |
| 156 |
AT1G08310 |
AT1G08310
|
[alpha/beta-Hydrolases superfamily protein] |
-3.514 |
-1.352 |
| 157 |
AT2G37030 |
AT2G37030
|
SAUR46, SMALL AUXIN UPREGULATED RNA 46 |
-3.423 |
-2.823 |
| 158 |
AT5G12030 |
AT5G12030
|
AT-HSP17.6A, heat shock protein 17.6A, HSP17.6, HEAT SHOCK PROTEIN 17.6, HSP17.6A, heat shock protein 17.6A |
-3.348 |
-0.402 |
| 159 |
AT1G73010 |
AT1G73010
|
AtPPsPase1, pyrophosphate-specific phosphatase1, ATPS2, phosphate starvation-induced gene 2, PPsPase1, pyrophosphate-specific phosphatase1, PS2, phosphate starvation-induced gene 2 |
-3.272 |
-0.757 |
| 160 |
AT4G36700 |
AT4G36700
|
[RmlC-like cupins superfamily protein] |
-3.251 |
1.348 |
| 161 |
AT1G53480 |
AT1G53480
|
ATMRD1, ARABIDOPSIS MTO 1 RESPONDING DOWN 1, MRD1, MTO 1 RESPONDING DOWN 1 |
-3.217 |
0.061 |
| 162 |
AT3G22231 |
AT3G22231
|
PCC1, PATHOGEN AND CIRCADIAN CONTROLLED 1 |
-3.183 |
-0.857 |
| 163 |
AT3G53250 |
AT3G53250
|
SAUR57, SMALL AUXIN UPREGULATED RNA 57 |
-3.178 |
-1.062 |
| 164 |
AT2G02680 |
AT2G02680
|
[Cysteine/Histidine-rich C1 domain family protein] |
-3.154 |
0.413 |
| 165 |
AT3G19680 |
AT3G19680
|
unknown |
-3.087 |
0.254 |
| 166 |
AT2G02990 |
AT2G02990
|
ATRNS1, RIBONUCLEASE 1, RNS1, ribonuclease 1 |
-3.015 |
-2.239 |
| 167 |
AT3G09450 |
AT3G09450
|
unknown |
-2.949 |
1.294 |
| 168 |
AT4G37990 |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
-2.875 |
0.719 |
| 169 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
-2.816 |
0.455 |
| 170 |
AT1G21910 |
AT1G21910
|
DREB26, dehydration response element-binding protein 26 |
-2.810 |
-0.882 |
| 171 |
AT1G25560 |
AT1G25560
|
AtTEM1, EDF1, ETHYLENE RESPONSE DNA BINDING FACTOR 1, TEM1, TEMPRANILLO 1 |
-2.795 |
-2.043 |
| 172 |
AT5G09570 |
AT5G09570
|
[Cox19-like CHCH family protein] |
-2.733 |
0.623 |
| 173 |
AT5G03545 |
AT5G03545
|
AT4, ATIPS2, INDUCED BY PI STARVATION 2 |
-2.704 |
-0.549 |
| 174 |
AT2G41240 |
AT2G41240
|
BHLH100, basic helix-loop-helix protein 100 |
-2.671 |
0.649 |
| 175 |
AT2G20670 |
AT2G20670
|
unknown |
-2.632 |
-0.734 |
| 176 |
AT1G13260 |
AT1G13260
|
EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR 4, RAV1, related to ABI3/VP1 1 |
-2.615 |
-0.504 |
| 177 |
AT3G59900 |
AT3G59900
|
ARGOS, AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE |
-2.560 |
-2.265 |
| 178 |
AT3G56970 |
AT3G56970
|
BHLH038, bHLH38, basic helix-loop-helix 38, ORG2, OBP3-RESPONSIVE GENE 3 |
-2.464 |
0.518 |
| 179 |
AT3G22235 |
AT3G22235
|
unknown |
-2.460 |
-1.147 |
| 180 |
AT3G56980 |
AT3G56980
|
BHLH039, bHLH39, basic helix-loop-helix 39, ORG3, OBP3-RESPONSIVE GENE 3 |
-2.420 |
0.036 |
| 181 |
AT5G04150 |
AT5G04150
|
BHLH101 |
-2.399 |
0.279 |
| 182 |
AT5G56550 |
AT5G56550
|
ATOXS3, OXIDATIVE STRESS 3, OXS3, OXIDATIVE STRESS 3 |
-2.394 |
-0.685 |
| 183 |
AT1G77765 |
AT1G77765
|
unknown |
-2.391 |
-0.519 |
| 184 |
AT5G40820 |
AT5G40820
|
ATATR, ARABIDOPSIS THALIANA ATAXIA TELANGIECTASIA-MUTATED AND RAD3-RELATED, ATR, Ataxia telangiectasia-mutated and RAD3-related, ATRAD3 |
-2.375 |
-0.250 |
| 185 |
AT2G14560 |
AT2G14560
|
LURP1, LATE UPREGULATED IN RESPONSE TO HYALOPERONOSPORA PARASITICA |
-2.365 |
-0.854 |
| 186 |
AT2G15020 |
AT2G15020
|
unknown |
-2.363 |
0.999 |
| 187 |
AT1G80440 |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-2.319 |
-1.448 |
| 188 |
AT2G40330 |
AT2G40330
|
PYL6, PYR1-like 6, RCAR9, regulatory components of ABA receptor 9 |
-2.307 |
-3.440 |
| 189 |
AT5G17300 |
AT5G17300
|
RVE1, REVEILLE 1 |
-2.307 |
0.057 |
| 190 |
AT1G74290 |
AT1G74290
|
[alpha/beta-Hydrolases superfamily protein] |
-2.276 |
0.246 |
| 191 |
AT4G08950 |
AT4G08950
|
EXO, EXORDIUM |
-2.267 |
0.798 |
| 192 |
AT5G44260 |
AT5G44260
|
AtTZF5, TZF5, Tandem CCCH Zinc Finger protein 5 |
-2.243 |
-1.163 |
| 193 |
AT1G64360 |
AT1G64360
|
unknown |
-2.230 |
-1.035 |
| 194 |
AT2G44080 |
AT2G44080
|
ARL, ARGOS-like |
-2.218 |
-2.174 |
| 195 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-2.175 |
0.266 |
| 196 |
AT1G72430 |
AT1G72430
|
SAUR78, SMALL AUXIN UPREGULATED RNA 78 |
-2.154 |
-0.884 |
| 197 |
AT1G49210 |
AT1G49210
|
[RING/U-box superfamily protein] |
-2.139 |
-2.631 |
| 198 |
AT5G05440 |
AT5G05440
|
PYL5, PYRABACTIN RESISTANCE 1-LIKE 5, RCAR8, regulatory component of ABA receptor 8 |
-2.131 |
-1.480 |
| 199 |
AT1G50040 |
AT1G50040
|
unknown |
-2.121 |
0.728 |
| 200 |
AT1G23140 |
AT1G23140
|
[Calcium-dependent lipid-binding (CaLB domain) family protein] |
-2.114 |
-0.592 |
| 201 |
AT2G23290 |
AT2G23290
|
AtMYB70, myb domain protein 70, MYB70, myb domain protein 70 |
-2.094 |
0.321 |
| 202 |
AT3G11110 |
AT3G11110
|
[RING/U-box superfamily protein] |
-2.081 |
0.862 |
| 203 |
AT4G12320 |
AT4G12320
|
CYP706A6, cytochrome P450, family 706, subfamily A, polypeptide 6 |
-2.070 |
0.768 |
| 204 |
AT4G20820 |
AT4G20820
|
[FAD-binding Berberine family protein] |
-2.067 |
0.902 |
| 205 |
AT1G52342 |
AT1G52342
|
unknown |
-2.063 |
0.095 |
| 206 |
AT4G34770 |
AT4G34770
|
SAUR1, SMALL AUXIN UPREGULATED RNA 1 |
-2.046 |
-6.438 |
| 207 |
AT5G26220 |
AT5G26220
|
AtGGCT2;1, GGCT2;1, gamma-glutamyl cyclotransferase 2;1 |
-2.033 |
1.102 |
| 208 |
AT1G02610 |
AT1G02610
|
[RING/FYVE/PHD zinc finger superfamily protein] |
-2.005 |
-3.754 |
| 209 |
AT5G23350 |
AT5G23350
|
[GRAM domain-containing protein / ABA-responsive protein-related] |
-2.003 |
-0.169 |
| 210 |
AT1G23390 |
AT1G23390
|
[Kelch repeat-containing F-box family protein] |
-1.998 |
-0.284 |
| 211 |
AT3G47420 |
AT3G47420
|
AtG3Pp1, Glycerol-3-phosphate permease 1, ATPS3, phosphate starvation-induced gene 3, G3Pp1, Glycerol-3-phosphate permease 1, PS3, phosphate starvation-induced gene 3 |
-1.995 |
-3.443 |
| 212 |
AT2G44460 |
AT2G44460
|
BGLU28, beta glucosidase 28 |
-1.988 |
-0.509 |
| 213 |
AT1G35140 |
AT1G35140
|
EXL1, EXORDIUM like 1, PHI-1, PHOSPHATE-INDUCED 1 |
-1.971 |
2.169 |
| 214 |
AT1G03870 |
AT1G03870
|
FLA9, FASCICLIN-like arabinoogalactan 9 |
-1.966 |
-0.918 |
| 215 |
AT3G04070 |
AT3G04070
|
anac047, NAC domain containing protein 47, NAC047, NAC domain containing protein 47 |
-1.964 |
-1.734 |
| 216 |
AT3G05630 |
AT3G05630
|
PDLZ2, PHOSPHOLIPASE D ZETA 2, PLDP2, phospholipase D P2, PLDZETA2, PHOSPHOLIPASE D ZETA 2 |
-1.957 |
-2.225 |
| 217 |
AT2G41640 |
AT2G41640
|
[Glycosyltransferase family 61 protein] |
-1.949 |
0.038 |
| 218 |
AT5G04190 |
AT5G04190
|
PKS4, phytochrome kinase substrate 4 |
-1.947 |
-5.315 |
| 219 |
AT3G26320 |
AT3G26320
|
CYP71B36, cytochrome P450, family 71, subfamily B, polypeptide 36 |
-1.923 |
-0.369 |
| 220 |
AT1G72416 |
AT1G72416
|
[Chaperone DnaJ-domain superfamily protein] |
-1.916 |
-4.156 |
| 221 |
AT5G19190 |
AT5G19190
|
unknown |
-1.913 |
-0.378 |
| 222 |
AT5G62360 |
AT5G62360
|
[Plant invertase/pectin methylesterase inhibitor superfamily protein] |
-1.903 |
1.670 |
| 223 |
AT1G73830 |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-1.888 |
-0.441 |
| 224 |
AT5G02760 |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-1.884 |
-2.991 |
| 225 |
AT1G06160 |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
-1.873 |
-3.036 |
| 226 |
AT2G32290 |
AT2G32290
|
BAM6, beta-amylase 6, BMY5, BETA-AMYLASE 5 |
-1.872 |
-0.186 |
| 227 |
AT1G79770 |
AT1G79770
|
unknown |
-1.864 |
-0.802 |
| 228 |
AT5G61590 |
AT5G61590
|
[Integrase-type DNA-binding superfamily protein] |
-1.853 |
-2.920 |
| 229 |
AT5G35777 |
AT5G35777
|
[copia-like retrotransposon family, has a 2.0e-67 P-value blast match to GB:CAA72989 open reading frame 1 (Ty1_Copia-element) (Brassica oleracea)] |
-1.852 |
-1.243 |
| 230 |
AT5G20150 |
AT5G20150
|
ATSPX1, ARABIDOPSIS THALIANA SPX DOMAIN GENE 1, SPX1, SPX domain gene 1 |
-1.840 |
-3.191 |
| 231 |
AT2G30230 |
AT2G30230
|
unknown |
-1.807 |
-0.231 |
| 232 |
AT1G29460 |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-1.806 |
-1.210 |
| 233 |
AT5G49740 |
AT5G49740
|
ATFRO7, FERRIC REDUCTION OXIDASE 7, FRO7, ferric reduction oxidase 7 |
-1.803 |
-0.275 |
| 234 |
AT1G12030 |
AT1G12030
|
unknown |
-1.769 |
1.980 |
| 235 |
AT1G18400 |
AT1G18400
|
BEE1, BR enhanced expression 1 |
-1.753 |
-3.908 |
| 236 |
AT3G22540 |
AT3G22540
|
unknown |
-1.741 |
0.550 |
| 237 |
AT1G68840 |
AT1G68840
|
AtRAV2, EDF2, ETHYLENE RESPONSE DNA BINDING FACTOR 2, RAP2.8, RELATED TO AP2 8, RAV2, related to ABI3/VP1 2, TEM2, TEMPRANILLO 2 |
-1.708 |
-0.515 |
| 238 |
AT1G66540 |
AT1G66540
|
[Cytochrome P450 superfamily protein] |
-1.700 |
0.394 |
| 239 |
AT1G12940 |
AT1G12940
|
ATNRT2.5, nitrate transporter2.5, NRT2.5, nitrate transporter2.5 |
-1.682 |
0.061 |
| 240 |
AT5G62280 |
AT5G62280
|
unknown |
-1.676 |
-3.215 |
| 241 |
AT2G34010 |
AT2G34010
|
TIE4, TCP Interactor containing EAR motif protein 4 |
-1.663 |
-0.615 |
| 242 |
AT1G78450 |
AT1G78450
|
[SOUL heme-binding family protein] |
-1.656 |
-1.017 |
| 243 |
AT1G75490 |
AT1G75490
|
[Integrase-type DNA-binding superfamily protein] |
-1.653 |
0.125 |
| 244 |
AT5G67060 |
AT5G67060
|
HEC1, HECATE 1 |
-1.651 |
-3.354 |
| 245 |
AT4G25780 |
AT4G25780
|
[CAP (Cysteine-rich secretory proteins, Antigen 5, and Pathogenesis-related 1 protein) superfamily protein] |
-1.647 |
-0.149 |
| 246 |
AT4G15550 |
AT4G15550
|
IAGLU, indole-3-acetate beta-D-glucosyltransferase |
-1.643 |
0.761 |
| 247 |
AT1G13670 |
AT1G13670
|
unknown |
-1.642 |
-2.628 |
| 248 |
AT2G47880 |
AT2G47880
|
[Glutaredoxin family protein] |
-1.642 |
-0.010 |
| 249 |
AT2G32100 |
AT2G32100
|
ATOFP16, RABIDOPSIS THALIANA OVATE FAMILY PROTEIN 16, OFP16, ovate family protein 16 |
-1.636 |
-5.170 |
| 250 |
AT4G17460 |
AT4G17460
|
HAT1, JAB, JAIBA |
-1.636 |
-1.761 |
| 251 |
AT4G12490 |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
-1.632 |
0.654 |
| 252 |
AT5G35732 |
AT5G35732
|
unknown |
-1.629 |
-2.493 |
| 253 |
AT5G18670 |
AT5G18670
|
BAM9, BETA-AMYLASE 9, BMY3, beta-amylase 3 |
-1.624 |
-0.331 |
| 254 |
AT4G35060 |
AT4G35060
|
HIPP25, heavy metal associated isoprenylated plant protein 25 |
-1.608 |
0.235 |
| 255 |
AT3G23880 |
AT3G23880
|
[F-box and associated interaction domains-containing protein] |
-1.603 |
-2.002 |
| 256 |
AT3G56200 |
AT3G56200
|
[Transmembrane amino acid transporter family protein] |
-1.600 |
0.690 |
| 257 |
AT2G30040 |
AT2G30040
|
MAPKKK14, mitogen-activated protein kinase kinase kinase 14 |
-1.597 |
-2.907 |
| 258 |
AT2G25200 |
AT2G25200
|
unknown |
-1.591 |
0.210 |
| 259 |
AT3G59940 |
AT3G59940
|
KFB50, Kelch repeat F-box 50, KMD4, KISS ME DEADLY 4 |
-1.585 |
-0.035 |
| 260 |
AT4G28290 |
AT4G28290
|
unknown |
-1.570 |
-1.240 |
| 261 |
AT3G06070 |
AT3G06070
|
unknown |
-1.556 |
-2.348 |
| 262 |
AT4G16563 |
AT4G16563
|
[Eukaryotic aspartyl protease family protein] |
-1.554 |
0.645 |
| 263 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-1.530 |
-0.338 |
| 264 |
AT4G37260 |
AT4G37260
|
ATMYB73, MYB73, myb domain protein 73 |
-1.517 |
0.694 |
| 265 |
AT5G21940 |
AT5G21940
|
unknown |
-1.515 |
-0.562 |
| 266 |
AT4G32480 |
AT4G32480
|
unknown |
-1.508 |
0.421 |
| 267 |
AT1G31173 |
AT1G31173
|
MIR167D, microRNA167D |
-1.507 |
-0.620 |
| 268 |
AT1G14430 |
AT1G14430
|
[glyoxal oxidase-related protein] |
-1.505 |
-1.146 |
| 269 |
AT1G73600 |
AT1G73600
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-1.499 |
0.068 |
| 270 |
AT4G37240 |
AT4G37240
|
unknown |
-1.495 |
0.791 |
| 271 |
AT2G01850 |
AT2G01850
|
ATXTH27, EXGT-A3, endoxyloglucan transferase A3, XTH27, XYLOGLUCAN ENDOTRANSGLUCOSYLASE/HYDROLASE 27 |
-1.494 |
-0.566 |
| 272 |
AT1G71030 |
AT1G71030
|
ATMYBL2, ARABIDOPSIS MYB-LIKE 2, MYBL2, MYB-like 2 |
-1.491 |
-0.860 |
| 273 |
AT3G48310 |
AT3G48310
|
CYP71A22, cytochrome P450, family 71, subfamily A, polypeptide 22 |
-1.488 |
-0.750 |
| 274 |
AT3G45860 |
AT3G45860
|
CRK4, cysteine-rich RLK (RECEPTOR-like protein kinase) 4 |
-1.484 |
-0.360 |
| 275 |
AT3G58120 |
AT3G58120
|
ATBZIP61, BZIP61 |
-1.462 |
-1.287 |
| 276 |
AT5G14730 |
AT5G14730
|
unknown |
-1.460 |
-2.761 |
| 277 |
AT3G01990 |
AT3G01990
|
ACR6, ACT domain repeat 6 |
-1.460 |
-1.319 |
| 278 |
AT5G45820 |
AT5G45820
|
CIPK20, CBL-interacting protein kinase 20, PKS18, PROTEIN KINASE 18, SnRK3.6, SNF1-RELATED PROTEIN KINASE 3.6 |
-1.460 |
0.029 |
| 279 |
AT3G06145 |
AT3G06145
|
unknown |
-1.457 |
-0.886 |
| 280 |
AT4G27310 |
AT4G27310
|
BBX28, B-box domain protein 28 |
-1.457 |
-0.432 |
| 281 |
AT2G38310 |
AT2G38310
|
PYL4, PYR1-like 4, RCAR10, regulatory components of ABA receptor 10 |
-1.456 |
-1.549 |
| 282 |
AT5G56840 |
AT5G56840
|
[myb-like transcription factor family protein] |
-1.454 |
-0.749 |
| 283 |
AT1G49200 |
AT1G49200
|
[RING/U-box superfamily protein] |
-1.441 |
-2.092 |
| 284 |
AT2G47440 |
AT2G47440
|
[Tetratricopeptide repeat (TPR)-like superfamily protein] |
-1.439 |
0.284 |
| 285 |
AT2G01890 |
AT2G01890
|
ATPAP8, PURPLE ACID PHOSPHATASE 8, PAP8, purple acid phosphatase 8 |
-1.438 |
-0.303 |
| 286 |
AT1G10020 |
AT1G10020
|
unknown |
-1.436 |
0.789 |
| 287 |
AT1G70985 |
AT1G70985
|
[hydroxyproline-rich glycoprotein family protein] |
-1.434 |
-1.415 |
| 288 |
AT1G23030 |
AT1G23030
|
[ARM repeat superfamily protein] |
-1.433 |
-0.147 |
| 289 |
AT1G66230 |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-1.432 |
-3.802 |
| 290 |
AT4G10120 |
AT4G10120
|
ATSPS4F |
-1.427 |
-0.789 |
| 291 |
AT2G30540 |
AT2G30540
|
[Thioredoxin superfamily protein] |
-1.418 |
-2.609 |
| 292 |
AT4G13810 |
AT4G13810
|
AtRLP47, receptor like protein 47, RLP47, receptor like protein 47 |
-1.411 |
-0.029 |
| 293 |
AT1G76530 |
AT1G76530
|
[Auxin efflux carrier family protein] |
-1.407 |
-0.032 |
| 294 |
AT5G19120 |
AT5G19120
|
[Eukaryotic aspartyl protease family protein] |
-1.395 |
-0.489 |
| 295 |
AT3G44450 |
AT3G44450
|
unknown |
-1.392 |
0.508 |
| 296 |
AT2G38940 |
AT2G38940
|
ATPT2, ARABIDOPSIS THALIANA PHOSPHATE TRANSPORTER 2, PHT1;4, phosphate transporter 1;4 |
-1.388 |
-0.154 |
| 297 |
AT1G18330 |
AT1G18330
|
EPR1, EARLY-PHYTOCHROME-RESPONSIVE1, RVE7, REVEILLE 7 |
-1.386 |
0.049 |