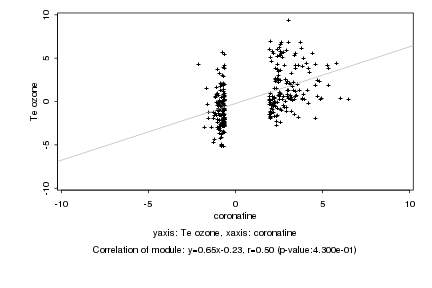
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
coronatine |
Log2 signal ratio
Te ozone |
| 1 |
AT5G07530 |
AT5G07530
|
ATGRP-7, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 7, ATGRP17, ARABIDOPSIS THALIANA GLYCINE RICH PROTEIN 17, GRP17, glycine rich protein 17 |
6.466 |
0.238 |
| 2 |
AT5G40350 |
AT5G40350
|
AtMYB24, myb domain protein 24, MYB24, myb domain protein 24 |
6.014 |
0.463 |
| 3 |
AT2G44578 |
AT2G44578
|
[RING/U-box superfamily protein] |
5.792 |
4.393 |
| 4 |
AT2G38240 |
AT2G38240
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
5.337 |
3.885 |
| 5 |
AT3G22275 |
AT3G22275
|
unknown |
5.328 |
1.883 |
| 6 |
AT1G30135 |
AT1G30135
|
JAZ8, jasmonate-zim-domain protein 8, TIFY5A |
5.242 |
4.163 |
| 7 |
AT5G40348 |
AT5G40348
|
other RNA |
4.942 |
0.462 |
| 8 |
AT1G12610 |
AT1G12610
|
DDF1, DWARF AND DELAYED FLOWERING 1 |
4.879 |
0.238 |
| 9 |
AT2G34600 |
AT2G34600
|
JAZ7, jasmonate-zim-domain protein 7, TIFY5B |
4.807 |
2.423 |
| 10 |
AT4G34410 |
AT4G34410
|
RRTF1, redox responsive transcription factor 1 |
4.706 |
2.433 |
| 11 |
AT4G17470 |
AT4G17470
|
[alpha/beta-Hydrolases superfamily protein] |
4.686 |
0.669 |
| 12 |
AT1G73325 |
AT1G73325
|
[Kunitz family trypsin and protease inhibitor protein] |
4.593 |
-1.873 |
| 13 |
AT5G63450 |
AT5G63450
|
CYP94B1, cytochrome P450, family 94, subfamily B, polypeptide 1 |
4.573 |
1.874 |
| 14 |
AT5G28237 |
AT5G28237
|
Pyridoxal-5'-phosphate-dependent enzyme family protein |
4.561 |
4.333 |
| 15 |
AT3G53600 |
AT3G53600
|
[C2H2-type zinc finger family protein] |
4.415 |
5.545 |
| 16 |
AT5G05600 |
AT5G05600
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
4.210 |
3.405 |
| 17 |
AT2G24210 |
AT2G24210
|
TPS10, terpene synthase 10 |
4.186 |
-0.141 |
| 18 |
AT4G36950 |
AT4G36950
|
MAPKKK21, mitogen-activated protein kinase kinase kinase 21 |
4.163 |
3.822 |
| 19 |
AT4G16740 |
AT4G16740
|
ATTPS03, terpene synthase 03, TPS03, terpene synthase 03 |
4.118 |
1.374 |
| 20 |
AT2G22760 |
AT2G22760
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
4.071 |
4.480 |
| 21 |
AT2G06045 |
AT2G06045
|
[copia-like retrotransposon family, has a 2.2e-12 P-value blast match to GB:AAC02672 polyprotein (Ty1_Copia-element) (Arabidopsis arenosa)] |
3.934 |
0.238 |
| 22 |
AT1G32910 |
AT1G32910
|
[HXXXD-type acyl-transferase family protein] |
3.886 |
0.915 |
| 23 |
AT1G17380 |
AT1G17380
|
JAZ5, jasmonate-zim-domain protein 5, TIFY11A |
3.867 |
5.046 |
| 24 |
AT5G45890 |
AT5G45890
|
AtSAG12, SAG12, senescence-associated gene 12 |
3.834 |
0.238 |
| 25 |
AT5G13220 |
AT5G13220
|
JAS1, JASMONATE-ASSOCIATED 1, JAZ10, jasmonate-zim-domain protein 10, TIFY9, TIFY DOMAIN PROTEIN 9 |
3.827 |
4.046 |
| 26 |
AT2G22770 |
AT2G22770
|
NAI1 |
3.809 |
0.443 |
| 27 |
AT1G43160 |
AT1G43160
|
RAP2.6, related to AP2 6 |
3.781 |
6.149 |
| 28 |
AT5G12340 |
AT5G12340
|
unknown |
3.713 |
6.900 |
| 29 |
AT3G48520 |
AT3G48520
|
CYP94B3, cytochrome P450, family 94, subfamily B, polypeptide 3 |
3.662 |
1.356 |
| 30 |
AT2G38750 |
AT2G38750
|
ANNAT4, annexin 4, AtANN4 |
3.614 |
-1.763 |
| 31 |
AT2G06050 |
AT2G06050
|
AtOPR3, DDE1, DELAYED DEHISCENCE 1, OPR3, oxophytodienoate-reductase 3 |
3.587 |
4.204 |
| 32 |
AT1G76640 |
AT1G76640
|
CML39, CALMODULIN LIKE 39 |
3.555 |
2.001 |
| 33 |
AT1G70700 |
AT1G70700
|
JAZ9, JASMONATE-ZIM-DOMAIN PROTEIN 9, TIFY7 |
3.483 |
0.769 |
| 34 |
AT1G11185 |
AT1G11185
|
other RNA |
3.442 |
4.160 |
| 35 |
AT1G32640 |
AT1G32640
|
ATMYC2, JAI1, JASMONATE INSENSITIVE 1, JIN1, JASMONATE INSENSITIVE 1, MYC2, RD22BP1, ZBF1 |
3.434 |
0.691 |
| 36 |
AT3G43250 |
AT3G43250
|
unknown |
3.426 |
5.552 |
| 37 |
AT1G19670 |
AT1G19670
|
ATCLH1, chlorophyllase 1, ATHCOR1, CORONATINE-INDUCED PROTEIN 1, CLH1, chlorophyllase 1, CORI1, CORONATINE-INDUCED PROTEIN 1 |
3.420 |
1.169 |
| 38 |
AT1G44350 |
AT1G44350
|
ILL6, IAA-leucine resistant (ILR)-like gene 6 |
3.416 |
3.900 |
| 39 |
AT3G25180 |
AT3G25180
|
CYP82G1, cytochrome P450, family 82, subfamily G, polypeptide 1 |
3.413 |
0.616 |
| 40 |
AT3G44860 |
AT3G44860
|
FAMT, farnesoic acid carboxyl-O-methyltransferase |
3.364 |
5.368 |
| 41 |
AT1G64195 |
AT1G64195
|
[Defensin-like (DEFL) family protein] |
3.355 |
0.238 |
| 42 |
AT2G38760 |
AT2G38760
|
ANN3, ANNEXIN 3, ANNAT3, annexin 3, AtANN3 |
3.336 |
-1.466 |
| 43 |
AT3G51450 |
AT3G51450
|
[Calcium-dependent phosphotriesterase superfamily protein] |
3.334 |
2.276 |
| 44 |
AT5G56880 |
AT5G56880
|
unknown |
3.294 |
1.375 |
| 45 |
AT3G25760 |
AT3G25760
|
AOC1, allene oxide cyclase 1, ERD12, early-responsive to dehydration 12 |
3.255 |
1.829 |
| 46 |
AT5G06870 |
AT5G06870
|
ATPGIP2, ARABIDOPSIS POLYGALACTURONASE INHIBITING PROTEIN 2, PGIP2, polygalacturonase inhibiting protein 2 |
3.250 |
0.118 |
| 47 |
AT1G51780 |
AT1G51780
|
ILL5, IAA-leucine resistant (ILR)-like gene 5 |
3.213 |
3.292 |
| 48 |
AT4G38000 |
AT4G38000
|
DOF4.7, DNA binding with one finger 4.7 |
3.206 |
0.507 |
| 49 |
AT1G18710 |
AT1G18710
|
AtMYB47, myb domain protein 47, MYB47, myb domain protein 47 |
3.205 |
-1.110 |
| 50 |
AT1G17420 |
AT1G17420
|
ATLOX3, Arabidopsis thaliana lipoxygenase 3, LOX3, lipoxygenase 3 |
3.125 |
0.747 |
| 51 |
AT4G27654 |
AT4G27654
|
unknown |
3.116 |
0.238 |
| 52 |
AT5G21960 |
AT5G21960
|
[Integrase-type DNA-binding superfamily protein] |
3.116 |
2.001 |
| 53 |
AT5G56980 |
AT5G56980
|
unknown |
3.048 |
2.188 |
| 54 |
AT5G43650 |
AT5G43650
|
BHLH92 |
3.041 |
6.907 |
| 55 |
AT5G67080 |
AT5G67080
|
MAPKKK19, mitogen-activated protein kinase kinase kinase 19 |
3.038 |
9.458 |
| 56 |
AT5G47240 |
AT5G47240
|
atnudt8, nudix hydrolase homolog 8, NUDT8, nudix hydrolase homolog 8 |
3.018 |
0.357 |
| 57 |
AT1G61890 |
AT1G61890
|
[MATE efflux family protein] |
3.010 |
1.378 |
| 58 |
AT1G74950 |
AT1G74950
|
JAZ2, JASMONATE-ZIM-DOMAIN PROTEIN 2, TIFY10B |
2.957 |
1.362 |
| 59 |
AT2G32150 |
AT2G32150
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
2.954 |
2.371 |
| 60 |
AT5G17490 |
AT5G17490
|
AtRGL3, RGL3, RGA-like protein 3 |
2.951 |
0.594 |
| 61 |
AT4G27140 |
AT4G27140
|
AT2S1, SESA1, seed storage albumin 1 |
2.948 |
0.916 |
| 62 |
AT1G66370 |
AT1G66370
|
AtMYB113, myb domain protein 113, MYB113, myb domain protein 113 |
2.922 |
2.166 |
| 63 |
AT2G32510 |
AT2G32510
|
MAPKKK17, mitogen-activated protein kinase kinase kinase 17 |
2.921 |
-0.646 |
| 64 |
AT3G15500 |
AT3G15500
|
ANAC055, NAC domain containing protein 55, ATNAC3, NAC domain containing protein 3, NAC055, NAC domain containing protein 55, NAC3, NAC domain containing protein 3 |
2.918 |
5.974 |
| 65 |
AT4G15440 |
AT4G15440
|
CYP74B2, HPL1, hydroperoxide lyase 1 |
2.901 |
-1.048 |
| 66 |
AT1G74930 |
AT1G74930
|
ORA47 |
2.858 |
0.053 |
| 67 |
AT1G06620 |
AT1G06620
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
2.830 |
2.606 |
| 68 |
AT1G52890 |
AT1G52890
|
ANAC019, NAC domain containing protein 19, NAC019, NAC domain containing protein 19 |
2.804 |
4.259 |
| 69 |
AT1G19550 |
AT1G19550
|
[Glutathione S-transferase family protein] |
2.765 |
-0.668 |
| 70 |
AT4G17500 |
AT4G17500
|
ATERF-1, ethylene responsive element binding factor 1, ERF-1, ethylene responsive element binding factor 1 |
2.742 |
5.673 |
| 71 |
AT5G67210 |
AT5G67210
|
IRX15-L, IRX15-LIKE |
2.712 |
0.655 |
| 72 |
AT5G19100 |
AT5G19100
|
[Eukaryotic aspartyl protease family protein] |
2.703 |
-0.441 |
| 73 |
AT4G29690 |
AT4G29690
|
[Alkaline-phosphatase-like family protein] |
2.668 |
0.263 |
| 74 |
AT1G51760 |
AT1G51760
|
IAR3, IAA-ALANINE RESISTANT 3, JR3, JASMONIC ACID RESPONSIVE 3 |
2.662 |
5.112 |
| 75 |
AT5G38120 |
AT5G38120
|
4CL8 |
2.634 |
-1.010 |
| 76 |
AT1G19180 |
AT1G19180
|
AtJAZ1, JAZ1, jasmonate-zim-domain protein 1, TIFY10A |
2.600 |
5.636 |
| 77 |
AT1G28480 |
AT1G28480
|
GRX480, roxy19 |
2.596 |
6.902 |
| 78 |
AT5G10300 |
AT5G10300
|
AtHNL, ATMES5, ARABIDOPSIS THALIANA METHYL ESTERASE 5, HNL, HYDROXYNITRILE LYASE, MES5, methyl esterase 5 |
2.581 |
0.914 |
| 79 |
AT5G59580 |
AT5G59580
|
UGT76E1, UDP-glucosyl transferase 76E1 |
2.570 |
1.021 |
| 80 |
AT4G24350 |
AT4G24350
|
[Phosphorylase superfamily protein] |
2.560 |
2.435 |
| 81 |
AT5G50760 |
AT5G50760
|
SAUR55, SMALL AUXIN UPREGULATED RNA 55 |
2.560 |
3.678 |
| 82 |
AT4G01080 |
AT4G01080
|
TBL26, TRICHOME BIREFRINGENCE-LIKE 26 |
2.552 |
-2.399 |
| 83 |
AT5G02940 |
AT5G02940
|
unknown |
2.542 |
-0.919 |
| 84 |
AT1G72520 |
AT1G72520
|
ATLOX4, Arabidopsis thaliana lipoxygenase 4, LOX4, lipoxygenase 4 |
2.540 |
5.844 |
| 85 |
AT5G47220 |
AT5G47220
|
ATERF-2, ETHYLENE RESPONSE FACTOR- 2, ATERF2, ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 2, ERF2, ethylene responsive element binding factor 2 |
2.538 |
5.372 |
| 86 |
AT3G23240 |
AT3G23240
|
ATERF1, ETHYLENE RESPONSE FACTOR 1, ERF1, ethylene response factor 1 |
2.532 |
6.588 |
| 87 |
AT4G10390 |
AT4G10390
|
[Protein kinase superfamily protein] |
2.508 |
0.309 |
| 88 |
AT4G24340 |
AT4G24340
|
[Phosphorylase superfamily protein] |
2.505 |
5.310 |
| 89 |
AT3G25780 |
AT3G25780
|
AOC3, allene oxide cyclase 3 |
2.494 |
6.290 |
| 90 |
AT1G79310 |
AT1G79310
|
AtMC7, metacaspase 7, AtMCP2a, metacaspase 2a, MC7, metacaspase 7, MCP2a, metacaspase 2a |
2.492 |
0.670 |
| 91 |
AT3G59750 |
AT3G59750
|
LecRK-V.8, L-type lectin receptor kinase V.8 |
2.492 |
0.859 |
| 92 |
AT3G18777 |
AT3G18777
|
[pseudogene] |
2.482 |
1.145 |
| 93 |
AT2G44840 |
AT2G44840
|
ATERF13, ETHYLENE-RESPONSIVE ELEMENT BINDING FACTOR 13, EREBP, ERF13, ethylene-responsive element binding factor 13 |
2.470 |
2.201 |
| 94 |
AT2G44070 |
AT2G44070
|
[NagB/RpiA/CoA transferase-like superfamily protein] |
2.468 |
0.787 |
| 95 |
AT5G53750 |
AT5G53750
|
[CBS domain-containing protein] |
2.462 |
2.586 |
| 96 |
AT2G39420 |
AT2G39420
|
[alpha/beta-Hydrolases superfamily protein] |
2.454 |
3.489 |
| 97 |
AT3G55970 |
AT3G55970
|
ATJRG21, JRG21, jasmonate-regulated gene 21 |
2.420 |
3.097 |
| 98 |
AT1G16370 |
AT1G16370
|
ATOCT6, ARABIDOPSIS THALIANA ORGANIC CATION/CARNITINE TRANSPORTER 6, OCT6, organic cation/carnitine transporter 6 |
2.412 |
3.772 |
| 99 |
AT5G24770 |
AT5G24770
|
ATVSP2, VSP2, vegetative storage protein 2 |
2.409 |
-0.745 |
| 100 |
AT1G20515 |
AT1G20515
|
other RNA |
2.408 |
0.001 |
| 101 |
AT4G16590 |
AT4G16590
|
ATCSLA01, cellulose synthase-like A01, ATCSLA1, CELLULOSE SYNTHASE-LIKE A1, CSLA01, CSLA01, cellulose synthase-like A01 |
2.406 |
-1.498 |
| 102 |
AT5G10625 |
AT5G10625
|
unknown |
2.404 |
4.316 |
| 103 |
AT4G21830 |
AT4G21830
|
ATMSRB7, methionine sulfoxide reductase B7, MSRB7, methionine sulfoxide reductase B7 |
2.378 |
6.056 |
| 104 |
AT1G61120 |
AT1G61120
|
GES, GERANYLLINALOOL SYNTHASE, TPS04, terpene synthase 04, TPS4, TERPENE SYNTHASE 4 |
2.371 |
5.243 |
| 105 |
AT1G72450 |
AT1G72450
|
JAZ6, jasmonate-zim-domain protein 6, TIFY11B, TIFY DOMAIN PROTEIN 11B |
2.364 |
2.744 |
| 106 |
AT1G61610 |
AT1G61610
|
[S-locus lectin protein kinase family protein] |
2.360 |
5.308 |
| 107 |
AT3G46700 |
AT3G46700
|
[UDP-Glycosyltransferase superfamily protein] |
2.337 |
2.511 |
| 108 |
AT4G02360 |
AT4G02360
|
unknown |
2.336 |
1.525 |
| 109 |
AT1G25422 |
AT1G25422
|
unknown |
2.336 |
-1.627 |
| 110 |
AT4G04840 |
AT4G04840
|
ATMSRB6, methionine sulfoxide reductase B6, MSRB6, methionine sulfoxide reductase B6 |
2.328 |
-2.225 |
| 111 |
AT5G38130 |
AT5G38130
|
[HXXXD-type acyl-transferase family protein] |
2.316 |
-2.750 |
| 112 |
AT4G27410 |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
2.289 |
2.329 |
| 113 |
AT3G45140 |
AT3G45140
|
ATLOX2, ARABIODOPSIS THALIANA LIPOXYGENASE 2, LOX2, lipoxygenase 2 |
2.287 |
-0.492 |
| 114 |
AT3G50280 |
AT3G50280
|
[HXXXD-type acyl-transferase family protein] |
2.284 |
2.676 |
| 115 |
AT1G64200 |
AT1G64200
|
VHA-E3, vacuolar H+-ATPase subunit E isoform 3 |
2.280 |
-0.078 |
| 116 |
AT2G27310 |
AT2G27310
|
[F-box family protein] |
2.258 |
3.896 |
| 117 |
AT1G20510 |
AT1G20510
|
OPCL1, OPC-8:0 CoA ligase1 |
2.247 |
2.566 |
| 118 |
AT3G25770 |
AT3G25770
|
AOC2, allene oxide cyclase 2 |
2.236 |
-0.359 |
| 119 |
AT5G52320 |
AT5G52320
|
CYP96A4, cytochrome P450, family 96, subfamily A, polypeptide 4 |
2.236 |
-1.715 |
| 120 |
AT4G15210 |
AT4G15210
|
AT-BETA-AMY, ATBETA-AMY, ARABIDOPSIS THALIANA BETA-AMYLASE, BAM5, beta-amylase 5, BMY1, RAM1, REDUCED BETA AMYLASE 1 |
2.224 |
-0.214 |
| 121 |
AT5G06865 |
AT5G06865
|
other RNA |
2.224 |
0.092 |
| 122 |
AT2G29450 |
AT2G29450
|
AT103-1A, ATGSTU1, ARABIDOPSIS THALIANA GLUTATHIONE S-TRANSFERASE TAU 1, ATGSTU5, glutathione S-transferase tau 5, GSTU5, glutathione S-transferase tau 5 |
2.213 |
0.349 |
| 123 |
AT3G06490 |
AT3G06490
|
AtMYB108, myb domain protein 108, BOS1, BOTRYTIS-SUSCEPTIBLE1, MYB108, myb domain protein 108 |
2.211 |
1.534 |
| 124 |
AT4G14680 |
AT4G14680
|
APS3 |
2.193 |
0.499 |
| 125 |
AT1G76790 |
AT1G76790
|
IGMT5, indole glucosinolate O-methyltransferase 5 |
2.184 |
-0.223 |
| 126 |
AT2G30100 |
AT2G30100
|
[pentatricopeptide (PPR) repeat-containing protein] |
2.171 |
-1.196 |
| 127 |
AT2G32130 |
AT2G32130
|
unknown |
2.166 |
5.547 |
| 128 |
AT1G74430 |
AT1G74430
|
ATMYB95, ARABIDOPSIS THALIANA MYB DOMAIN PROTEIN 95, ATMYBCP66, ARABIDOPSIS THALIANA MYB DOMAIN CONTAINING PROTEIN 66, MYB95, myb domain protein 95 |
2.132 |
-0.463 |
| 129 |
AT5G59310 |
AT5G59310
|
LTP4, lipid transfer protein 4 |
2.123 |
0.581 |
| 130 |
AT2G45930 |
AT2G45930
|
unknown |
2.121 |
0.238 |
| 131 |
AT2G36590 |
AT2G36590
|
ATPROT3, PROLINE TRANSPORTER 3, ProT3, proline transporter 3 |
2.120 |
-0.389 |
| 132 |
AT1G06160 |
AT1G06160
|
ORA59, octadecanoid-responsive Arabidopsis AP2/ERF 59 |
2.099 |
5.841 |
| 133 |
AT5G53048 |
AT5G53048
|
other RNA |
2.073 |
-0.881 |
| 134 |
AT5G63160 |
AT5G63160
|
BT1, BTB and TAZ domain protein 1 |
2.061 |
-0.372 |
| 135 |
AT2G43530 |
AT2G43530
|
[Scorpion toxin-like knottin superfamily protein] |
2.053 |
-0.725 |
| 136 |
AT2G15760 |
AT2G15760
|
unknown |
2.046 |
4.725 |
| 137 |
AT1G12240 |
AT1G12240
|
ATBETAFRUCT4, AtFRUCT4, AtVI2, FRUCT4, fructosidase 4, VAC-INV, VACUOLAR INVERTASE, VI2, vacuolar invertase 2 |
2.020 |
-1.336 |
| 138 |
AT3G16690 |
AT3G16690
|
AtSWEET16, SWEET16 |
1.997 |
-1.095 |
| 139 |
AT1G65890 |
AT1G65890
|
AAE12, acyl activating enzyme 12 |
1.989 |
-1.920 |
| 140 |
AT5G19110 |
AT5G19110
|
[Eukaryotic aspartyl protease family protein] |
1.986 |
1.023 |
| 141 |
AT5G38710 |
AT5G38710
|
[Methylenetetrahydrofolate reductase family protein] |
1.985 |
6.990 |
| 142 |
AT1G56650 |
AT1G56650
|
ATMYB75, MYB DOMAIN PROTEIN 75, MYB75, MYELOBLASTOSIS PROTEIN 75, PAP1, production of anthocyanin pigment 1, SIAA1, SUC-INDUCED ANTHOCYANIN ACCUMULATION 1 |
1.983 |
-1.621 |
| 143 |
AT2G22930 |
AT2G22930
|
[UDP-Glycosyltransferase superfamily protein] |
1.981 |
-1.222 |
| 144 |
AT1G49530 |
AT1G49530
|
GGPS6, geranylgeranyl pyrophosphate synthase 6 |
1.979 |
0.170 |
| 145 |
AT2G22200 |
AT2G22200
|
[Integrase-type DNA-binding superfamily protein] |
1.977 |
-1.741 |
| 146 |
AT2G39030 |
AT2G39030
|
NATA1, N-acetyltransferase activity 1 |
1.968 |
5.087 |
| 147 |
AT5G42650 |
AT5G42650
|
AOS, allene oxide synthase, CYP74A, CYTOCHROME P450 74A, DDE2, DELAYED DEHISCENCE 2 |
1.963 |
-0.041 |
| 148 |
AT4G37410 |
AT4G37410
|
CYP81F4, cytochrome P450, family 81, subfamily F, polypeptide 4 |
1.958 |
-0.019 |
| 149 |
AT5G59330 |
AT5G59330
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
1.954 |
0.165 |
| 150 |
AT1G31550 |
AT1G31550
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
1.951 |
-1.488 |
| 151 |
AT4G37990 |
AT4G37990
|
ATCAD8, ARABIDOPSIS THALIANA CINNAMYL-ALCOHOL DEHYDROGENASE 8, CAD-B2, CINNAMYL-ALCOHOL DEHYDROGENASE B2, ELI3, ELICITOR-ACTIVATED GENE 3, ELI3-2, elicitor-activated gene 3-2 |
1.937 |
6.008 |
| 152 |
AT1G07050 |
AT1G07050
|
[CCT motif family protein] |
1.937 |
0.242 |
| 153 |
AT1G75960 |
AT1G75960
|
[AMP-dependent synthetase and ligase family protein] |
1.933 |
-0.320 |
| 154 |
AT4G05100 |
AT4G05100
|
AtMYB74, myb domain protein 74, MYB74, myb domain protein 74 |
1.922 |
0.636 |
| 155 |
AT3G55150 |
AT3G55150
|
ATEXO70H1, exocyst subunit exo70 family protein H1, EXO70H1, exocyst subunit exo70 family protein H1 |
-2.173 |
4.320 |
| 156 |
AT5G41900 |
AT5G41900
|
[alpha/beta-Hydrolases superfamily protein] |
-1.826 |
-2.955 |
| 157 |
AT1G29860 |
AT1G29860
|
ATWRKY71, WRKY DNA-BINDING PROTEIN 71, WRKY71, WRKY DNA-binding protein 71 |
-1.697 |
1.502 |
| 158 |
AT4G37890 |
AT4G37890
|
EDA40, embryo sac development arrest 40 |
-1.609 |
-0.306 |
| 159 |
AT4G01975 |
AT4G01975
|
[pseudogene] |
-1.570 |
-1.849 |
| 160 |
AT4G25470 |
AT4G25470
|
ATCBF2, CBF2, C-repeat/DRE binding factor 2, DREB1C, DRE/CRT-BINDING PROTEIN 1C, FTQ4, FREEZING TOLERANCE QTL 4 |
-1.563 |
-1.197 |
| 161 |
AT3G18217 |
AT3G18217
|
MIR157C, microRNA157C |
-1.427 |
-2.916 |
| 162 |
AT5G62730 |
AT5G62730
|
[Major facilitator superfamily protein] |
-1.313 |
-4.659 |
| 163 |
AT2G41170 |
AT2G41170
|
[F-box family protein] |
-1.313 |
-1.938 |
| 164 |
AT3G25905 |
AT3G25905
|
CLE27, CLAVATA3/ESR-RELATED 27 |
-1.295 |
-1.157 |
| 165 |
AT5G18080 |
AT5G18080
|
SAUR24, small auxin up RNA 24 |
-1.253 |
-4.318 |
| 166 |
AT1G78170 |
AT1G78170
|
unknown |
-1.222 |
-1.555 |
| 167 |
ATCG00400 |
ATCG00400
|
TRNL.1 |
-1.198 |
0.616 |
| 168 |
AT2G17470 |
AT2G17470
|
ALMT6, ALuminium activated Malate Transporter 6, AtALMT6 |
-1.194 |
-1.289 |
| 169 |
AT1G08115 |
AT1G08115
|
[gi|22293581|emb|AJ505685.1|ATH505685 Arabidopsis putative U1a snRNA (U1asnRNA gene), 5' end incomplete, At5g05684] |
-1.179 |
0.558 |
| 170 |
AT5G49330 |
AT5G49330
|
ATMYB111, ARABIDOPSIS MYB DOMAIN PROTEIN 111, MYB111, myb domain protein 111, PFG3, PRODUCTION OF FLAVONOL GLYCOSIDES 3 |
-1.145 |
-1.567 |
| 171 |
AT4G06477 |
AT4G06477
|
[gypsy-like retrotransposon family, has a 2.1e-112 P-value blast match to GB:AAD19359 polyprotein (gypsy_Ty3-element) (Sorghum bicolor)] |
-1.130 |
-0.020 |
| 172 |
AT4G21500 |
AT4G21500
|
unknown |
-1.128 |
1.719 |
| 173 |
AT2G28630 |
AT2G28630
|
KCS12, 3-ketoacyl-CoA synthase 12 |
-1.100 |
0.897 |
| 174 |
AT5G51850 |
AT5G51850
|
TRM24, TON1 Recruiting Motif 24 |
-1.084 |
-2.316 |
| 175 |
AT5G67450 |
AT5G67450
|
AZF1, zinc-finger protein 1, ZF1, zinc-finger protein 1 |
-1.074 |
3.719 |
| 176 |
AT5G36790 |
AT5G36790
|
[Haloacid dehalogenase-like hydrolase (HAD) superfamily protein] |
-1.065 |
-0.986 |
| 177 |
AT5G02760 |
AT5G02760
|
APD7, Arabidopsis Pp2c clade D 7 |
-1.054 |
-2.964 |
| 178 |
AT4G32950 |
AT4G32950
|
[Protein phosphatase 2C family protein] |
-1.038 |
-0.413 |
| 179 |
AT4G02075 |
AT4G02075
|
PIT1, pitchoun 1 |
-1.037 |
1.029 |
| 180 |
AT4G23870 |
AT4G23870
|
unknown |
-1.037 |
-2.063 |
| 181 |
AT3G30180 |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-1.023 |
-0.876 |
| 182 |
AT3G23930 |
AT3G23930
|
unknown |
-1.013 |
-1.323 |
| 183 |
AT1G15405 |
AT1G15405
|
other RNA |
-1.010 |
-0.311 |
| 184 |
AT5G47610 |
AT5G47610
|
[RING/U-box superfamily protein] |
-1.010 |
-3.161 |
| 185 |
AT5G26010 |
AT5G26010
|
[Protein phosphatase 2C family protein] |
-1.006 |
-0.860 |
| 186 |
AT4G20820 |
AT4G20820
|
[FAD-binding Berberine family protein] |
-1.001 |
-1.003 |
| 187 |
ATCG00570 |
ATCG00570
|
PSBF, photosystem II reaction center protein F |
-0.999 |
0.762 |
| 188 |
AT1G77870 |
AT1G77870
|
MUB5, membrane-anchored ubiquitin-fold protein 5 precursor |
-0.990 |
-0.745 |
| 189 |
ATCG00580 |
ATCG00580
|
PSBE, photosystem II reaction center protein E |
-0.978 |
-0.179 |
| 190 |
AT1G49200 |
AT1G49200
|
[RING/U-box superfamily protein] |
-0.972 |
-0.955 |
| 191 |
AT1G19840 |
AT1G19840
|
SAUR53, SMALL AUXIN UPREGULATED RNA 53 |
-0.970 |
0.079 |
| 192 |
AT2G46970 |
AT2G46970
|
PIL1, phytochrome interacting factor 3-like 1 |
-0.960 |
-1.007 |
| 193 |
AT5G24660 |
AT5G24660
|
LSU2, RESPONSE TO LOW SULFUR 2 |
-0.959 |
-0.415 |
| 194 |
ATCG00450 |
ATCG00450
|
TRNV.1 |
-0.949 |
0.151 |
| 195 |
AT5G54148 |
AT5G54148
|
unknown |
-0.946 |
0.097 |
| 196 |
AT5G54145 |
AT5G54145
|
unknown |
-0.946 |
-2.076 |
| 197 |
AT3G09450 |
AT3G09450
|
unknown |
-0.945 |
-2.845 |
| 198 |
AT5G62280 |
AT5G62280
|
unknown |
-0.942 |
-3.004 |
| 199 |
AT1G02390 |
AT1G02390
|
ATGPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2, GPAT2, GLYCEROL-3-PHOSPHATE sn-2-ACYLTRANSFERASE 2 |
-0.940 |
3.346 |
| 200 |
AT4G32280 |
AT4G32280
|
IAA29, indole-3-acetic acid inducible 29 |
-0.936 |
-3.618 |
| 201 |
AT5G25240 |
AT5G25240
|
unknown |
-0.933 |
-1.592 |
| 202 |
AT5G55620 |
AT5G55620
|
unknown |
-0.920 |
-2.088 |
| 203 |
AT5G10930 |
AT5G10930
|
CIPK5, CBL-interacting protein kinase 5, SnRK3.24, SNF1-RELATED PROTEIN KINASE 3.24 |
-0.916 |
-2.166 |
| 204 |
AT5G12050 |
AT5G12050
|
unknown |
-0.903 |
-2.828 |
| 205 |
AT2G30424 |
AT2G30424
|
TCL2, TRICHOMELESS 2 |
-0.899 |
-1.606 |
| 206 |
AT5G53730 |
AT5G53730
|
NHL26, NDR1/HIN1-like 26 |
-0.898 |
-0.706 |
| 207 |
AT1G29440 |
AT1G29440
|
SAUR63, SMALL AUXIN UP RNA 63 |
-0.889 |
-4.180 |
| 208 |
AT1G73830 |
AT1G73830
|
BEE3, BR enhanced expression 3 |
-0.889 |
1.770 |
| 209 |
AT4G18830 |
AT4G18830
|
ATOFP5, OFP5, ovate family protein 5 |
-0.887 |
-0.565 |
| 210 |
AT3G25670 |
AT3G25670
|
[Leucine-rich repeat (LRR) family protein] |
-0.884 |
-3.298 |
| 211 |
AT5G45340 |
AT5G45340
|
CYP707A3, cytochrome P450, family 707, subfamily A, polypeptide 3 |
-0.877 |
2.167 |
| 212 |
AT5G03700 |
AT5G03700
|
[D-mannose binding lectin protein with Apple-like carbohydrate-binding domain] |
-0.871 |
1.376 |
| 213 |
AT1G11700 |
AT1G11700
|
unknown |
-0.865 |
-2.563 |
| 214 |
AT2G02950 |
AT2G02950
|
PKS1, phytochrome kinase substrate 1 |
-0.859 |
-0.565 |
| 215 |
AT2G40610 |
AT2G40610
|
ATEXP8, ATEXPA8, expansin A8, ATHEXP ALPHA 1.11, EXP8, EXPA8, expansin A8 |
-0.857 |
-4.095 |
| 216 |
AT5G18030 |
AT5G18030
|
[SAUR-like auxin-responsive protein family ] |
-0.844 |
-5.093 |
| 217 |
AT5G18020 |
AT5G18020
|
SAUR20, SMALL AUXIN UP RNA 20 |
-0.841 |
-4.896 |
| 218 |
AT4G38620 |
AT4G38620
|
ATMYB4, myb domain protein 4, MYB4, myb domain protein 4 |
-0.835 |
-0.055 |
| 219 |
AT2G24762 |
AT2G24762
|
AtGDU4, glutamine dumper 4, GDU4, glutamine dumper 4 |
-0.830 |
-2.516 |
| 220 |
AT2G01670 |
AT2G01670
|
atnudt17, nudix hydrolase homolog 17, NUDT17, nudix hydrolase homolog 17 |
-0.828 |
2.032 |
| 221 |
AT1G02380 |
AT1G02380
|
unknown |
-0.826 |
-1.508 |
| 222 |
AT1G29500 |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
-0.815 |
-5.055 |
| 223 |
ATCG01100 |
ATCG01100
|
NDHA |
-0.812 |
0.467 |
| 224 |
AT1G29510 |
AT1G29510
|
SAUR67, SMALL AUXIN UPREGULATED RNA 67 |
-0.810 |
-3.697 |
| 225 |
AT1G04220 |
AT1G04220
|
KCS2, 3-ketoacyl-CoA synthase 2 |
-0.809 |
-0.548 |
| 226 |
AT1G49000 |
AT1G49000
|
unknown |
-0.787 |
5.674 |
| 227 |
AT2G33060 |
AT2G33060
|
AtRLP27, receptor like protein 27, RLP27, receptor like protein 27 |
-0.779 |
-0.730 |
| 228 |
AT2G36800 |
AT2G36800
|
DOGT1, don-glucosyltransferase 1, UGT73C5, UDP-GLUCOSYL TRANSFERASE 73C5 |
-0.777 |
1.996 |
| 229 |
AT1G29450 |
AT1G29450
|
SAUR64, SMALL AUXIN UPREGULATED RNA 64 |
-0.776 |
-3.627 |
| 230 |
AT2G18890 |
AT2G18890
|
[Protein kinase superfamily protein] |
-0.775 |
-2.414 |
| 231 |
AT5G25190 |
AT5G25190
|
ESE3, ethylene and salt inducible 3 |
-0.770 |
-1.090 |
| 232 |
AT5G03995 |
AT5G03995
|
unknown |
-0.769 |
-0.168 |
| 233 |
AT1G67050 |
AT1G67050
|
unknown |
-0.767 |
-0.728 |
| 234 |
AT4G39070 |
AT4G39070
|
BBX20, B-box domain protein 20, BZS1, BZS1 |
-0.764 |
3.003 |
| 235 |
AT1G15260 |
AT1G15260
|
unknown |
-0.759 |
-2.661 |
| 236 |
AT5G45790 |
AT5G45790
|
[Ubiquitin carboxyl-terminal hydrolase family protein] |
-0.758 |
-0.677 |
| 237 |
AT3G03850 |
AT3G03850
|
SAUR26, SMALL AUXIN UP RNA 26 |
-0.757 |
-2.825 |
| 238 |
AT3G41768 |
AT3G41768
|
[18SrRNA] |
-0.753 |
-0.002 |
| 239 |
AT5G37300 |
AT5G37300
|
WSD1 |
-0.746 |
-2.027 |
| 240 |
AT1G79110 |
AT1G79110
|
BRG2, BOI-related gene 2 |
-0.745 |
0.059 |
| 241 |
AT4G01680 |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-0.743 |
0.628 |
| 242 |
AT2G17040 |
AT2G17040
|
anac036, NAC domain containing protein 36, NAC036, NAC domain containing protein 36 |
-0.742 |
1.435 |
| 243 |
AT4G29905 |
AT4G29905
|
unknown |
-0.742 |
-3.398 |
| 244 |
AT4G34970 |
AT4G34970
|
ADF9, actin depolymerizing factor 9 |
-0.739 |
0.464 |
| 245 |
AT3G61950 |
AT3G61950
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.735 |
-2.097 |
| 246 |
AT1G13080 |
AT1G13080
|
CYP71B2, cytochrome P450, family 71, subfamily B, polypeptide 2 |
-0.734 |
-1.915 |
| 247 |
AT2G40130 |
AT2G40130
|
SMXL8, SMAX1-like 8 |
-0.733 |
-2.484 |
| 248 |
AT1G69160 |
AT1G69160
|
unknown |
-0.726 |
-1.526 |
| 249 |
AT1G24530 |
AT1G24530
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.726 |
2.905 |
| 250 |
AT4G04745 |
AT4G04745
|
unknown |
-0.721 |
0.883 |
| 251 |
AT1G29460 |
AT1G29460
|
SAUR65, SMALL AUXIN UPREGULATED RNA 65 |
-0.721 |
-5.137 |
| 252 |
AT4G14400 |
AT4G14400
|
ACD6, ACCELERATED CELL DEATH 6 |
-0.718 |
-1.326 |
| 253 |
AT2G47485 |
AT2G47485
|
unknown |
-0.718 |
0.688 |
| 254 |
AT5G61340 |
AT5G61340
|
unknown |
-0.718 |
-1.581 |
| 255 |
AT4G23880 |
AT4G23880
|
unknown |
-0.717 |
-0.295 |
| 256 |
AT3G60580 |
AT3G60580
|
[C2H2-like zinc finger protein] |
-0.717 |
1.229 |
| 257 |
AT5G53500 |
AT5G53500
|
[Transducin/WD40 repeat-like superfamily protein] |
-0.716 |
-1.578 |
| 258 |
AT2G41990 |
AT2G41990
|
unknown |
-0.715 |
-0.825 |
| 259 |
AT5G43700 |
AT5G43700
|
ATAUX2-11, AUXIN INDUCIBLE 2-11, IAA4, indole-3-acetic acid inducible 4 |
-0.715 |
-1.751 |
| 260 |
ATCG00140 |
ATCG00140
|
ATPH |
-0.712 |
0.008 |
| 261 |
AT5G34800 |
AT5G34800
|
[pseudogene] |
-0.710 |
-2.386 |
| 262 |
AT1G70420 |
AT1G70420
|
unknown |
-0.708 |
0.149 |
| 263 |
AT5G41761 |
AT5G41761
|
unknown |
-0.708 |
2.132 |
| 264 |
AT2G12462 |
AT2G12462
|
unknown |
-0.708 |
-1.410 |
| 265 |
AT3G15570 |
AT3G15570
|
[Phototropic-responsive NPH3 family protein] |
-0.707 |
-2.838 |
| 266 |
AT4G23020 |
AT4G23020
|
TRM11, TON1 Recruiting Motif 11 |
-0.707 |
-2.191 |
| 267 |
AT5G34790 |
AT5G34790
|
[CACTA-like transposase family (Ptta/En/Spm), has a 2.9e-11 P-value blast match to At5g36655.1/81-333 CACTA-like transposase family (Ptta/En/Spm) (CACTA-element) (Arabidopsis thaliana)] |
-0.702 |
-1.922 |
| 268 |
AT3G03820 |
AT3G03820
|
SAUR29, SMALL AUXIN UP RNA 29 |
-0.700 |
-5.161 |
| 269 |
AT1G13100 |
AT1G13100
|
CYP71B29, cytochrome P450, family 71, subfamily B, polypeptide 29 |
-0.699 |
-0.228 |
| 270 |
AT1G80440 |
AT1G80440
|
KFB20, Kelch repeat F-box 20, KMD1, KISS ME DEADLY 1 |
-0.698 |
1.440 |
| 271 |
AT5G52750 |
AT5G52750
|
[Heavy metal transport/detoxification superfamily protein ] |
-0.698 |
3.997 |
| 272 |
AT5G35732 |
AT5G35732
|
unknown |
-0.694 |
0.595 |
| 273 |
AT1G69490 |
AT1G69490
|
ANAC029, Arabidopsis NAC domain containing protein 29, ATNAP, NAC-LIKE, ACTIVATED BY AP3/PI, NAP, NAC-like, activated by AP3/PI |
-0.693 |
1.921 |
| 274 |
AT3G15310 |
AT3G15310
|
unknown |
-0.692 |
-2.544 |
| 275 |
AT3G59880 |
AT3G59880
|
unknown |
-0.685 |
3.812 |
| 276 |
AT5G11070 |
AT5G11070
|
unknown |
-0.684 |
-1.463 |
| 277 |
AT4G01460 |
AT4G01460
|
[basic helix-loop-helix (bHLH) DNA-binding superfamily protein] |
-0.681 |
-1.978 |
| 278 |
AT4G36470 |
AT4G36470
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-0.681 |
-1.024 |
| 279 |
AT2G43340 |
AT2G43340
|
unknown |
-0.681 |
-1.827 |
| 280 |
AT4G22730 |
AT4G22730
|
[Leucine-rich repeat protein kinase family protein] |
-0.679 |
-1.994 |
| 281 |
AT5G57780 |
AT5G57780
|
P1R1, P1R1 |
-0.678 |
-2.057 |
| 282 |
AT1G17990 |
AT1G17990
|
[FMN-linked oxidoreductases superfamily protein] |
-0.676 |
1.016 |
| 283 |
AT4G23810 |
AT4G23810
|
ATWRKY53, WRKY53 |
-0.676 |
5.458 |
| 284 |
AT3G62550 |
AT3G62550
|
[Adenine nucleotide alpha hydrolases-like superfamily protein] |
-0.673 |
-1.965 |
| 285 |
AT2G43290 |
AT2G43290
|
MSS3, multicopy suppressors of snf4 deficiency in yeast 3 |
-0.673 |
2.039 |
| 286 |
AT1G22250 |
AT1G22250
|
unknown |
-0.671 |
0.659 |
| 287 |
AT5G44680 |
AT5G44680
|
[DNA glycosylase superfamily protein] |
-0.671 |
-2.329 |
| 288 |
AT4G26540 |
AT4G26540
|
[Leucine-rich repeat receptor-like protein kinase family protein] |
-0.671 |
-2.879 |
| 289 |
ATCG00020 |
ATCG00020
|
PSBA, photosystem II reaction center protein A |
-0.670 |
0.217 |
| 290 |
AT4G34135 |
AT4G34135
|
UGT73B2, UDP-glucosyltransferase 73B2 |
-0.670 |
4.231 |
| 291 |
AT3G46130 |
AT3G46130
|
ATMYB48, myb domain protein 48, ATMYB48-1, ATMYB48-2, ATMYB48-3, MYB48, myb domain protein 48 |
-0.670 |
-2.026 |
| 292 |
AT5G47370 |
AT5G47370
|
HAT2 |
-0.670 |
-2.490 |
| 293 |
AT3G25717 |
AT3G25717
|
DVL6, DEVIL 6, RTFL16, ROTUNDIFOLIA like 16 |
-0.670 |
-2.745 |
| 294 |
AT4G38860 |
AT4G38860
|
SAUR16, SMALL AUXIN UPREGULATED RNA 16 |
-0.667 |
-3.517 |
| 295 |
AT3G23010 |
AT3G23010
|
AtRLP36, receptor like protein 36, RLP36, receptor like protein 36 |
-0.664 |
0.238 |
| 296 |
AT1G77020 |
AT1G77020
|
[DNAJ heat shock N-terminal domain-containing protein] |
-0.664 |
-1.012 |
| 297 |
AT5G18290 |
AT5G18290
|
SIP1;2, SIP1B, SMALL AND BASIC INTRINSIC PROTEIN 1B |
-0.662 |
0.208 |
| 298 |
AT1G14280 |
AT1G14280
|
PKS2, phytochrome kinase substrate 2 |
-0.661 |
-2.264 |
| 299 |
AT5G62470 |
AT5G62470
|
ATMYB96, MYB DOMAIN PROTEIN 96, MYB96, myb domain protein 96, MYBCOV1 |
-0.660 |
-0.362 |
| 300 |
AT5G49665 |
AT5G49665
|
[Zinc finger (C3HC4-type RING finger) family protein] |
-0.657 |
-0.909 |
| 301 |
ATCG00330 |
ATCG00330
|
RPS14, chloroplast ribosomal protein S14 |
-0.656 |
0.286 |
| 302 |
ATCG01130 |
ATCG01130
|
TIC214, TRANSLOCON AT THE INNER ENVELOPE MEMBRANE OF CHLOROPLASTS 214, YCF1.2 |
-0.654 |
-0.726 |
| 303 |
AT3G02380 |
AT3G02380
|
ATCOL2, CONSTANS-LIKE 2, BBX3, B-box domain protein 3, COL2, CONSTANS-like 2 |
-0.652 |
-2.582 |
| 304 |
AT1G66230 |
AT1G66230
|
AtMYB20, myb domain protein 20, MYB20, myb domain protein 20 |
-0.651 |
0.695 |
| 305 |
AT3G17640 |
AT3G17640
|
[Leucine-rich repeat (LRR) family protein] |
-0.647 |
-2.691 |
| 306 |
AT1G34640 |
AT1G34640
|
[peptidases] |
-0.646 |
-1.845 |
| 307 |
AT3G26932 |
AT3G26932
|
DRB3, dsRNA-binding protein 3 |
-0.645 |
-1.743 |