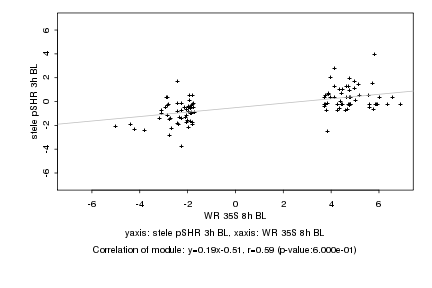
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
WR 35S 8h BL |
Log2 signal ratio
stele pSHR 3h BL |
| 1 |
AT2G23270 |
AT2G23270
|
unknown |
6.883 |
-0.169 |
| 2 |
AT5G20630 |
AT5G20630
|
ATGER3, ARABIDOPSIS THALIANA GERMIN 3, GER3, germin 3, GLP3, GERMIN-LIKE PROTEIN 3, GLP3A, GLP3B |
6.539 |
0.358 |
| 3 |
AT2G04050 |
AT2G04050
|
[MATE efflux family protein] |
6.342 |
-0.169 |
| 4 |
AT1G26380 |
AT1G26380
|
[FAD-binding Berberine family protein] |
6.011 |
0.378 |
| 5 |
AT1G31580 |
AT1G31580
|
CXC750, ECS1 |
5.917 |
-0.169 |
| 6 |
AT3G16660 |
AT3G16660
|
[Pollen Ole e 1 allergen and extensin family protein] |
5.863 |
-0.169 |
| 7 |
AT3G16670 |
AT3G16670
|
[Pollen Ole e 1 allergen and extensin family protein] |
5.832 |
-0.169 |
| 8 |
AT3G05730 |
AT3G05730
|
[LOCATED IN: endomembrane system] |
5.790 |
3.970 |
| 9 |
AT5G38430 |
AT5G38430
|
RBCS1B, Rubisco small subunit 1B |
5.731 |
-0.592 |
| 10 |
AT3G14420 |
AT3G14420
|
GOX1, glycolate oxidase 1 |
5.726 |
1.513 |
| 11 |
AT2G04070 |
AT2G04070
|
[MATE efflux family protein] |
5.575 |
-0.169 |
| 12 |
AT2G30750 |
AT2G30750
|
CYP71A12, cytochrome P450, family 71, subfamily A, polypeptide 12 |
5.567 |
-0.431 |
| 13 |
AT5G38420 |
AT5G38420
|
RBCS2B, Rubisco small subunit 2B |
5.521 |
0.526 |
| 14 |
AT5G43450 |
AT5G43450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
5.172 |
0.513 |
| 15 |
AT2G10940 |
AT2G10940
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
5.139 |
1.512 |
| 16 |
AT3G47340 |
AT3G47340
|
ASN1, glutamine-dependent asparagine synthase 1, AT-ASN1, ARABIDOPSIS THALIANA GLUTAMINE-DEPENDENT ASPARAGINE SYNTHASE 1, DIN6, DARK INDUCIBLE 6 |
5.011 |
0.166 |
| 17 |
AT5G38410 |
AT5G38410
|
RBCS3B, Rubisco small subunit 3B |
4.974 |
1.721 |
| 18 |
AT4G26260 |
AT4G26260
|
MIOX4, myo-inositol oxygenase 4 |
4.956 |
1.143 |
| 19 |
AT1G26390 |
AT1G26390
|
[FAD-binding Berberine family protein] |
4.798 |
-0.169 |
| 20 |
AT2G36800 |
AT2G36800
|
DOGT1, don-glucosyltransferase 1, UGT73C5, UDP-GLUCOSYL TRANSFERASE 73C5 |
4.795 |
0.358 |
| 21 |
AT4G17090 |
AT4G17090
|
AtBAM3, BAM3, BETA-AMYLASE 3, BMY8, BETA-AMYLASE 8, CT-BMY, chloroplast beta-amylase |
4.774 |
-0.183 |
| 22 |
AT2G34430 |
AT2G34430
|
LHB1B1, light-harvesting chlorophyll-protein complex II subunit B1, LHCB1.4, LIGHT-HARVESTING CHLOROPHYLL-PROTEIN COMPLEX II SUBUNIT B1 |
4.753 |
0.979 |
| 23 |
AT1G19150 |
AT1G19150
|
LHCA2*1, Lhca6, photosystem I light harvesting complex gene 6 |
4.751 |
0.358 |
| 24 |
AT5G09570 |
AT5G09570
|
[Cox19-like CHCH family protein] |
4.749 |
2.009 |
| 25 |
AT3G23250 |
AT3G23250
|
ATMYB15, MYB DOMAIN PROTEIN 15, ATY19, MYB15, myb domain protein 15 |
4.748 |
-0.282 |
| 26 |
AT3G45060 |
AT3G45060
|
ATNRT2.6, ARABIDOPSIS THALIANA HIGH AFFINITY NITRATE TRANSPORTER 2.6, NRT2.6, high affinity nitrate transporter 2.6 |
4.727 |
-0.169 |
| 27 |
AT3G01500 |
AT3G01500
|
ATBCA1, BETA CARBONIC ANHYDRASE 1, ATSABP3, ARABIDOPSIS THALIANA SALICYLIC ACID-BINDING PROTEIN 3, CA1, carbonic anhydrase 1, SABP3, SALICYLIC ACID-BINDING PROTEIN 3 |
4.722 |
1.330 |
| 28 |
AT4G12500 |
AT4G12500
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
4.705 |
-0.169 |
| 29 |
AT2G05070 |
AT2G05070
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.2, photosystem II light harvesting complex gene 2.2 |
4.658 |
-0.653 |
| 30 |
AT1G23130 |
AT1G23130
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
4.632 |
1.299 |
| 31 |
AT5G24640 |
AT5G24640
|
unknown |
4.615 |
0.358 |
| 32 |
AT5G33355 |
AT5G33355
|
[Defensin-like (DEFL) family protein] |
4.594 |
-0.703 |
| 33 |
AT5G25260 |
AT5G25260
|
[SPFH/Band 7/PHB domain-containing membrane-associated protein family] |
4.447 |
-0.169 |
| 34 |
AT2G32690 |
AT2G32690
|
ATGRP23, GLYCINE-RICH PROTEIN 23, GRP23, glycine-rich protein 23 |
4.442 |
1.047 |
| 35 |
AT1G21500 |
AT1G21500
|
unknown |
4.433 |
0.743 |
| 36 |
AT4G12490 |
AT4G12490
|
[Bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin superfamily protein] |
4.400 |
-0.169 |
| 37 |
AT5G14740 |
AT5G14740
|
BETA CA2, BETA CARBONIC ANHYDRASE 2, CA18, CARBONIC ANHYDRASE 18, CA2, carbonic anhydrase 2 |
4.383 |
0.068 |
| 38 |
AT5G26000 |
AT5G26000
|
AtTGG1, BGLU38, BETA GLUCOSIDASE 38, TGG1, thioglucoside glucohydrolase 1 |
4.345 |
1.048 |
| 39 |
AT1G29920 |
AT1G29920
|
AB165, CAB2, chlorophyll A/B-binding protein 2, LHCB1.1, LIGHT HARVESTING CHLOROPHYLL A/B-BINDING PROTEIN 1.1 |
4.329 |
-0.518 |
| 40 |
AT1G66100 |
AT1G66100
|
[Plant thionin] |
4.247 |
-0.169 |
| 41 |
AT5G39120 |
AT5G39120
|
[RmlC-like cupins superfamily protein] |
4.247 |
-0.703 |
| 42 |
AT3G27690 |
AT3G27690
|
LHCB2, LIGHT-HARVESTING CHLOROPHYLL B-BINDING 2, LHCB2.3, photosystem II light harvesting complex gene 2.3, LHCB2.4 |
4.140 |
2.822 |
| 43 |
AT3G46780 |
AT3G46780
|
PTAC16, plastid transcriptionally active 16 |
4.133 |
0.358 |
| 44 |
AT1G55260 |
AT1G55260
|
LTPG6, glycosylphosphatidylinositol-anchored lipid protein transfer 6 |
4.109 |
1.298 |
| 45 |
AT2G21330 |
AT2G21330
|
AtFBA1, FBA1, fructose-bisphosphate aldolase 1 |
3.961 |
0.358 |
| 46 |
AT1G56250 |
AT1G56250
|
AtPP2-B14, phloem protein 2-B14, PP2-B14, phloem protein 2-B14, VBF, VIP1-binding F-box protein |
3.945 |
2.057 |
| 47 |
AT4G32950 |
AT4G32950
|
[Protein phosphatase 2C family protein] |
3.866 |
0.743 |
| 48 |
AT5G25610 |
AT5G25610
|
ATRD22, RD22, RESPONSIVE TO DESSICATION 22 |
3.851 |
0.634 |
| 49 |
AT1G29910 |
AT1G29910
|
AB180, CAB3, chlorophyll A/B binding protein 3, LHCB1.2, LIGHT HARVESTING CHLOROPHYLL A/B BINDING PROTEIN 1.2 |
3.845 |
-0.085 |
| 50 |
AT5G52390 |
AT5G52390
|
[PAR1 protein] |
3.822 |
-2.472 |
| 51 |
AT3G14520 |
AT3G14520
|
[Terpenoid cyclases/Protein prenyltransferases superfamily protein] |
3.798 |
-0.703 |
| 52 |
AT4G30540 |
AT4G30540
|
[Class I glutamine amidotransferase-like superfamily protein] |
3.763 |
-0.169 |
| 53 |
AT2G42220 |
AT2G42220
|
[Rhodanese/Cell cycle control phosphatase superfamily protein] |
3.755 |
0.567 |
| 54 |
AT5G46360 |
AT5G46360
|
ATKCO3, CA2+ ACTIVATED OUTWARD RECTIFYING K+ CHANNEL 3, KCO3, Ca2+ activated outward rectifying K+ channel 3 |
3.721 |
0.358 |
| 55 |
AT4G14060 |
AT4G14060
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
3.716 |
-0.169 |
| 56 |
AT1G47603 |
AT1G47603
|
ATPUP19, purine permease 19, PUP19, purine permease 19 |
3.702 |
-0.353 |
| 57 |
AT3G62760 |
AT3G62760
|
ATGSTF13 |
-5.037 |
-2.091 |
| 58 |
AT4G04930 |
AT4G04930
|
DES-1-LIKE |
-4.420 |
-1.865 |
| 59 |
AT5G38940 |
AT5G38940
|
[RmlC-like cupins superfamily protein] |
-4.253 |
-2.310 |
| 60 |
AT5G22580 |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-3.838 |
-2.423 |
| 61 |
AT1G76220 |
AT1G76220
|
unknown |
-3.203 |
-1.416 |
| 62 |
AT5G44582 |
AT5G44582
|
unknown |
-3.098 |
-0.936 |
| 63 |
AT5G48130 |
AT5G48130
|
[Phototropic-responsive NPH3 family protein] |
-3.093 |
-0.703 |
| 64 |
AT3G44870 |
AT3G44870
|
[S-adenosyl-L-methionine-dependent methyltransferases superfamily protein] |
-2.943 |
-0.429 |
| 65 |
AT3G09330 |
AT3G09330
|
[Transmembrane amino acid transporter family protein] |
-2.914 |
0.383 |
| 66 |
AT4G15360 |
AT4G15360
|
CYP705A3, cytochrome P450, family 705, subfamily A, polypeptide 3 |
-2.878 |
0.358 |
| 67 |
AT2G21510 |
AT2G21510
|
[DNAJ heat shock N-terminal domain-containing protein] |
-2.849 |
-0.304 |
| 68 |
AT5G07550 |
AT5G07550
|
ATGRP19, GRP19, glycine-rich protein 19 |
-2.841 |
-1.092 |
| 69 |
AT5G20260 |
AT5G20260
|
[Exostosin family protein] |
-2.823 |
-0.169 |
| 70 |
AT5G66080 |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-2.771 |
-2.816 |
| 71 |
AT5G62210 |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-2.761 |
-1.467 |
| 72 |
AT1G76620 |
AT1G76620
|
unknown |
-2.727 |
-1.398 |
| 73 |
AT5G57785 |
AT5G57785
|
unknown |
-2.675 |
-2.235 |
| 74 |
AT2G24850 |
AT2G24850
|
TAT, TYROSINE AMINOTRANSFERASE, TAT3, tyrosine aminotransferase 3 |
-2.449 |
1.697 |
| 75 |
AT3G49630 |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.436 |
-1.826 |
| 76 |
AT1G64930 |
AT1G64930
|
CYP89A7, cytochrome P450, family 87, subfamily A, polypeptide 7 |
-2.434 |
-0.117 |
| 77 |
AT4G33145 |
AT4G33145
|
unknown |
-2.425 |
-0.837 |
| 78 |
AT1G53130 |
AT1G53130
|
GRI, GRIM REAPER |
-2.405 |
-1.855 |
| 79 |
AT3G09280 |
AT3G09280
|
unknown |
-2.378 |
-1.337 |
| 80 |
AT4G01680 |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-2.274 |
-0.141 |
| 81 |
AT2G46850 |
AT2G46850
|
[Protein kinase superfamily protein] |
-2.267 |
-3.703 |
| 82 |
AT5G66800 |
AT5G66800
|
unknown |
-2.260 |
-1.352 |
| 83 |
AT3G58000 |
AT3G58000
|
[VQ motif-containing protein] |
-2.256 |
-0.703 |
| 84 |
AT5G07190 |
AT5G07190
|
ATS3, seed gene 3 |
-2.160 |
-0.476 |
| 85 |
AT4G27170 |
AT4G27170
|
AT2S4, SESA4, seed storage albumin 4 |
-2.123 |
-1.301 |
| 86 |
AT5G35940 |
AT5G35940
|
[Mannose-binding lectin superfamily protein] |
-2.087 |
-0.587 |
| 87 |
AT2G41990 |
AT2G41990
|
unknown |
-2.073 |
-1.125 |
| 88 |
AT1G26870 |
AT1G26870
|
ANAC009, Arabidopsis NAC domain containing protein 9, FEZ, FEZ |
-2.072 |
-1.690 |
| 89 |
AT3G25655 |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-2.024 |
-1.514 |
| 90 |
AT4G33800 |
AT4G33800
|
unknown |
-1.998 |
-2.178 |
| 91 |
AT2G26870 |
AT2G26870
|
NPC2, non-specific phospholipase C2 |
-1.982 |
-0.482 |
| 92 |
AT1G59940 |
AT1G59940
|
ARR3, response regulator 3 |
-1.980 |
-0.793 |
| 93 |
AT3G56220 |
AT3G56220
|
[transcription regulators] |
-1.974 |
-0.381 |
| 94 |
AT3G01840 |
AT3G01840
|
LYK2, LysM-containing receptor-like kinase 2 |
-1.957 |
0.569 |
| 95 |
AT1G33612 |
AT1G33612
|
[Leucine-rich repeat (LRR) family protein] |
-1.930 |
0.132 |
| 96 |
AT3G62280 |
AT3G62280
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
-1.915 |
-0.466 |
| 97 |
AT2G04480 |
AT2G04480
|
unknown |
-1.902 |
-1.000 |
| 98 |
AT5G10510 |
AT5G10510
|
AIL6, AINTEGUMENTA-like 6, PLT3, PLETHORA 3 |
-1.897 |
-1.641 |
| 99 |
AT3G59440 |
AT3G59440
|
[Calcium-binding EF-hand family protein] |
-1.884 |
-0.526 |
| 100 |
AT3G30180 |
AT3G30180
|
BR6OX2, brassinosteroid-6-oxidase 2, CYP85A2 |
-1.878 |
-0.922 |
| 101 |
AT2G23050 |
AT2G23050
|
MEL4, MAB4/ENP/NPY1-LIKE 4, NPY4, NAKED PINS IN YUC MUTANTS 4 |
-1.868 |
-0.498 |
| 102 |
AT2G17070 |
AT2G17070
|
unknown |
-1.857 |
-0.877 |
| 103 |
AT2G25130 |
AT2G25130
|
[ARM repeat superfamily protein] |
-1.856 |
-0.952 |
| 104 |
AT1G49110 |
AT1G49110
|
unknown |
-1.844 |
-0.278 |
| 105 |
AT3G52440 |
AT3G52440
|
[Dof-type zinc finger DNA-binding family protein] |
-1.831 |
-0.175 |
| 106 |
AT1G02575 |
AT1G02575
|
unknown |
-1.826 |
-1.884 |
| 107 |
AT1G05835 |
AT1G05835
|
[PHD finger protein] |
-1.825 |
-1.717 |
| 108 |
AT4G15370 |
AT4G15370
|
BARS1, baruol synthase 1, PEN2, PENTACYCLIC TRITERPENE SYNTHASE 2 |
-1.820 |
0.589 |
| 109 |
AT4G35200 |
AT4G35200
|
unknown |
-1.780 |
-0.429 |
| 110 |
AT5G52170 |
AT5G52170
|
HDG7, homeodomain GLABROUS 7 |
-1.769 |
-0.105 |
| 111 |
AT3G50870 |
AT3G50870
|
GATA18, GATA TRANSCRIPTION FACTOR 18, HAN, HANABA TANARU, MNP, MONOPOLE |
-1.745 |
-0.909 |