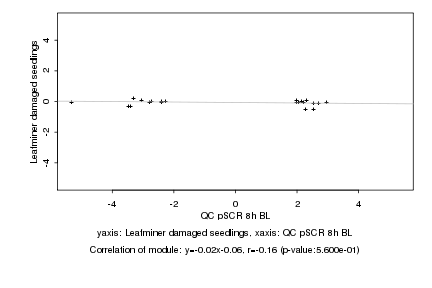
|
probeID |
AGICode |
Annotation |
Log2 signal ratio
QC pSCR 8h BL |
Log2 signal ratio
Leafminer damaged seedlings |
| 1 |
AT1G77950 |
AT1G77950
|
AGL67, AGAMOUS-like 67 |
2.951 |
-0.022 |
| 2 |
AT4G08620 |
AT4G08620
|
SULTR1;1, sulphate transporter 1;1 |
2.668 |
-0.082 |
| 3 |
AT1G29500 |
AT1G29500
|
SAUR66, SMALL AUXIN UPREGULATED RNA 66 |
2.526 |
-0.508 |
| 4 |
AT2G20570 |
AT2G20570
|
GBF's pro-rich region-interacting factor 1 |
2.510 |
-0.075 |
| 5 |
AT4G39675 |
AT4G39675
|
unknown |
2.290 |
0.080 |
| 6 |
AT1G26210 |
AT1G26210
|
ATSOFL1, SOB FIVE-LIKE 1, SOFL1, SOB five-like 1 |
2.257 |
-0.483 |
| 7 |
AT2G31540 |
AT2G31540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
2.228 |
no data |
| 8 |
AT5G04120 |
AT5G04120
|
[Phosphoglycerate mutase family protein] |
2.209 |
-0.043 |
| 9 |
AT4G06746 |
AT4G06746
|
DEAR5, DREB AND EAR MOTIF PROTEIN 5, RAP2.9, related to AP2 9 |
2.202 |
no data |
| 10 |
AT5G19890 |
AT5G19890
|
[Peroxidase superfamily protein] |
2.132 |
0.021 |
| 11 |
AT5G35390 |
AT5G35390
|
PRK1, pollen receptor like kinase 1 |
2.016 |
-0.053 |
| 12 |
AT1G73120 |
AT1G73120
|
unknown |
1.964 |
-0.043 |
| 13 |
AT3G05936 |
AT3G05936
|
unknown |
1.963 |
no data |
| 14 |
AT3G52820 |
AT3G52820
|
ATPAP22, PURPLE ACID PHOSPHATASE 22, PAP22, purple acid phosphatase 22 |
1.959 |
0.070 |
| 15 |
AT3G62760 |
AT3G62760
|
ATGSTF13 |
-5.349 |
-0.019 |
| 16 |
AT5G22580 |
AT5G22580
|
[Stress responsive A/B Barrel Domain] |
-3.491 |
-0.276 |
| 17 |
AT5G62210 |
AT5G62210
|
[Embryo-specific protein 3, (ATS3)] |
-3.413 |
-0.277 |
| 18 |
AT1G32540 |
AT1G32540
|
LOL1, lsd one like 1 |
-3.323 |
0.256 |
| 19 |
AT5G38940 |
AT5G38940
|
[RmlC-like cupins superfamily protein] |
-3.215 |
no data |
| 20 |
AT3G53232 |
AT3G53232
|
DVL20, DEVIL 20, RTFL1, ROTUNDIFOLIA like 1 |
-3.065 |
no data |
| 21 |
AT5G66080 |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-3.063 |
0.107 |
| 22 |
AT1G67623 |
AT1G67623
|
[F-box family protein] |
-2.805 |
-0.009 |
| 23 |
AT1G26870 |
AT1G26870
|
ANAC009, Arabidopsis NAC domain containing protein 9, FEZ, FEZ |
-2.758 |
0.020 |
| 24 |
AT1G03800 |
AT1G03800
|
ATERF10, ARABIDOPSIS THALIANA RF DOMAIN PROTEIN 10, ERF10, ERF domain protein 10 |
-2.554 |
no data |
| 25 |
AT3G49630 |
AT3G49630
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-2.425 |
-0.012 |
| 26 |
AT1G33612 |
AT1G33612
|
[Leucine-rich repeat (LRR) family protein] |
-2.410 |
0.040 |
| 27 |
AT3G25655 |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-2.279 |
0.066 |