|
probeID |
AGICode |
Annotation |
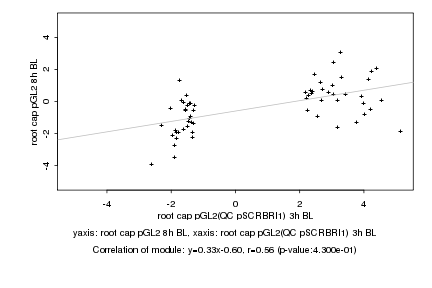
Log2 signal ratio
root cap pGL2(QC pSCRBRI1) 3h BL |
Log2 signal ratio
root cap pGL2 8h BL |
| 1 |
AT5G36925 |
AT5G36925
|
unknown |
5.126 |
-1.840 |
| 2 |
AT4G37780 |
AT4G37780
|
ATMYB87, MYB87, myb domain protein 87 |
4.516 |
0.079 |
| 3 |
AT1G52690 |
AT1G52690
|
LEA7, LATE EMBRYOGENESIS ABUNDANT 7 |
4.386 |
2.099 |
| 4 |
AT4G03480 |
AT4G03480
|
[Ankyrin repeat family protein] |
4.208 |
1.907 |
| 5 |
AT5G36920 |
AT5G36920
|
unknown |
4.177 |
-0.472 |
| 6 |
AT4G18195 |
AT4G18195
|
ATPUP8, ARABIDOPSIS THALIANA PURINE PERMEASE 8, PUP8, purine permease 8 |
4.132 |
1.437 |
| 7 |
AT5G05390 |
AT5G05390
|
LAC12, laccase 12 |
4.007 |
-0.749 |
| 8 |
AT5G48430 |
AT5G48430
|
[Eukaryotic aspartyl protease family protein] |
3.963 |
-0.071 |
| 9 |
AT4G14365 |
AT4G14365
|
XBAT34, XB3 ortholog 4 in Arabidopsis thaliana |
3.903 |
0.368 |
| 10 |
AT4G23170 |
AT4G23170
|
CRK9, CYSTEINE-RICH RLK (RECEPTOR-LIKE PROTEIN KINASE) 9, EP1 |
3.756 |
-1.287 |
| 11 |
AT2G26150 |
AT2G26150
|
ATHSFA2, heat shock transcription factor A2, HSFA2, heat shock transcription factor A2 |
3.404 |
0.467 |
| 12 |
AT1G51913 |
AT1G51913
|
[LOCATED IN: endomembrane system] |
3.289 |
1.528 |
| 13 |
AT1G22890 |
AT1G22890
|
unknown |
3.269 |
3.075 |
| 14 |
AT4G11480 |
AT4G11480
|
CRK32, cysteine-rich RLK (RECEPTOR-like protein kinase) 32 |
3.173 |
0.079 |
| 15 |
AT1G21320 |
AT1G21320
|
[nucleotide binding] |
3.146 |
-1.608 |
| 16 |
AT5G37490 |
AT5G37490
|
[ARM repeat superfamily protein] |
3.045 |
0.480 |
| 17 |
AT2G02010 |
AT2G02010
|
GAD4, glutamate decarboxylase 4 |
3.036 |
2.464 |
| 18 |
AT1G74710 |
AT1G74710
|
ATICS1, ARABIDOPSIS ISOCHORISMATE SYNTHASE 1, EDS16, ENHANCED DISEASE SUSCEPTIBILITY TO ERYSIPHE ORONTII 16, ICS1, ISOCHORISMATE SYNTHASE 1, SID2, SALICYLIC ACID INDUCTION DEFICIENT 2 |
3.011 |
1.045 |
| 19 |
AT1G22900 |
AT1G22900
|
[Disease resistance-responsive (dirigent-like protein) family protein] |
2.868 |
0.595 |
| 20 |
AT2G31540 |
AT2G31540
|
[GDSL-like Lipase/Acylhydrolase superfamily protein] |
2.681 |
0.752 |
| 21 |
AT4G14450 |
AT4G14450
|
unknown |
2.665 |
0.079 |
| 22 |
AT1G49000 |
AT1G49000
|
unknown |
2.636 |
1.238 |
| 23 |
AT5G20230 |
AT5G20230
|
ATBCB, blue-copper-binding protein, BCB, BLUE COPPER BINDING PROTEIN, BCB, blue-copper-binding protein, SAG14, SENESCENCE ASSOCIATED GENE 14 |
2.527 |
-0.905 |
| 24 |
AT1G14540 |
AT1G14540
|
PER4, peroxidase 4 |
2.444 |
1.718 |
| 25 |
AT4G04700 |
AT4G04700
|
CPK27, calcium-dependent protein kinase 27 |
2.382 |
0.677 |
| 26 |
AT4G15417 |
AT4G15417
|
ATRTL1, RNAse II-like 1, RTL1, RNAse II-like 1 |
2.348 |
0.525 |
| 27 |
AT4G21870 |
AT4G21870
|
[HSP20-like chaperones superfamily protein] |
2.307 |
0.719 |
| 28 |
AT3G50470 |
AT3G50470
|
HR3, homolog of RPW8 3, MLA10, INTRACELLULAR MILDEW A 10 |
2.264 |
0.413 |
| 29 |
AT5G64750 |
AT5G64750
|
ABR1, ABA REPRESSOR1 |
2.221 |
-0.551 |
| 30 |
AT1G07620 |
AT1G07620
|
ATOBGM, TRM32, TON1 Recruiting Motif 32 |
2.205 |
0.204 |
| 31 |
AT5G52640 |
AT5G52640
|
ATHS83, AtHsp90-1, HEAT SHOCK PROTEIN 90-1, ATHSP90.1, heat shock protein 90.1, HSP81-1, HEAT SHOCK PROTEIN 81-1, HSP81.1, HSP83, HEAT SHOCK PROTEIN 83, HSP90.1, heat shock protein 90.1 |
2.171 |
0.586 |
| 32 |
AT5G65300 |
AT5G65300
|
unknown |
-2.631 |
-3.909 |
| 33 |
AT1G63720 |
AT1G63720
|
unknown |
-2.303 |
-1.481 |
| 34 |
AT1G77760 |
AT1G77760
|
GNR1, NIA1, nitrate reductase 1, NR1, NITRATE REDUCTASE 1 |
-2.042 |
-0.408 |
| 35 |
AT3G49570 |
AT3G49570
|
LSU3, RESPONSE TO LOW SULFUR 3 |
-1.963 |
-2.110 |
| 36 |
AT4G01680 |
AT4G01680
|
AtMYB55, myb domain protein 55, MYB55, myb domain protein 55 |
-1.925 |
-3.453 |
| 37 |
AT3G15030 |
AT3G15030
|
MEE35, maternal effect embryo arrest 35, TCP4, TCP family transcription factor 4 |
-1.920 |
-2.748 |
| 38 |
AT2G25450 |
AT2G25450
|
[2-oxoglutarate (2OG) and Fe(II)-dependent oxygenase superfamily protein] |
-1.872 |
-1.761 |
| 39 |
AT3G25655 |
AT3G25655
|
IDL1, inflorescence deficient in abscission (IDA)-like 1 |
-1.856 |
-1.894 |
| 40 |
AT5G17800 |
AT5G17800
|
AtMYB56, myb domain protein 56, MYB56, myb domain protein 56 |
-1.855 |
-2.295 |
| 41 |
AT3G49580 |
AT3G49580
|
LSU1, RESPONSE TO LOW SULFUR 1 |
-1.774 |
-1.919 |
| 42 |
AT2G25090 |
AT2G25090
|
AtCIPK16, CIPK16, CBL-interacting protein kinase 16, SnRK3.18, SNF1-RELATED PROTEIN KINASE 3.18 |
-1.764 |
1.357 |
| 43 |
AT5G38200 |
AT5G38200
|
[Class I glutamine amidotransferase-like superfamily protein] |
-1.692 |
0.123 |
| 44 |
AT3G50660 |
AT3G50660
|
AtDWF4, CLM, CLOMAZONE-RESISTANT, CYP90B1, CYTOCHROME P450 90B1, DWF4, DWARF 4, PSC1, PARTIALLY SUPPRESSING COI1 INSENSITIVITY TO JA 1, SAV1, SHADE AVOIDANCE 1, SNP2, SUPPRESSOR OF NPH4 2 |
-1.629 |
-1.733 |
| 45 |
AT4G36220 |
AT4G36220
|
CYP84A1, CYTOCHROME P450 84A1, FAH1, ferulic acid 5-hydroxylase 1 |
-1.617 |
-0.031 |
| 46 |
AT5G65340 |
AT5G65340
|
unknown |
-1.572 |
-0.445 |
| 47 |
AT4G30270 |
AT4G30270
|
MERI-5, MERISTEM 5, MERI5B, meristem-5, SEN4, SENESCENCE 4, XTH24, xyloglucan endotransglucosylase/hydrolase 24 |
-1.560 |
-0.557 |
| 48 |
AT4G27410 |
AT4G27410
|
ANAC072, Arabidopsis NAC domain containing protein 72, RD26, RESPONSIVE TO DESICCATION 26 |
-1.534 |
0.433 |
| 49 |
AT2G35200 |
AT2G35200
|
unknown |
-1.510 |
-1.510 |
| 50 |
AT5G03390 |
AT5G03390
|
unknown |
-1.499 |
-0.210 |
| 51 |
AT3G23570 |
AT3G23570
|
[alpha/beta-Hydrolases superfamily protein] |
-1.474 |
-1.187 |
| 52 |
AT5G14200 |
AT5G14200
|
ATIMD1, ARABIDOPSIS ISOPROPYLMALATE DEHYDROGENASE 1, IMD1, isopropylmalate dehydrogenase 1 |
-1.452 |
-0.082 |
| 53 |
AT2G42430 |
AT2G42430
|
ASL18, ASYMMETRIC LEAVES2-LIKE 18, LBD16, lateral organ boundaries-domain 16 |
-1.441 |
-1.002 |
| 54 |
AT2G26370 |
AT2G26370
|
[MD-2-related lipid recognition domain-containing protein] |
-1.422 |
-0.887 |
| 55 |
AT4G38770 |
AT4G38770
|
ATPRP4, ARABIDOPSIS THALIANA PROLINE-RICH PROTEIN 4, PRP4, proline-rich protein 4 |
-1.410 |
-0.115 |
| 56 |
AT2G23620 |
AT2G23620
|
ATMES1, ARABIDOPSIS THALIANA METHYL ESTERASE 1, MES1, methyl esterase 1 |
-1.383 |
-1.259 |
| 57 |
AT3G13380 |
AT3G13380
|
BRL3, BRI1-like 3 |
-1.355 |
-1.890 |
| 58 |
AT5G66080 |
AT5G66080
|
APD9, Arabidopsis Pp2c clade D 9 |
-1.340 |
-2.217 |
| 59 |
AT1G51170 |
AT1G51170
|
AGC2-3, AGC2 kinase 3, UCN, UNICORN |
-1.319 |
-1.313 |
| 60 |
AT3G54600 |
AT3G54600
|
DJ-1f, DJ1F |
-1.313 |
-0.547 |
| 61 |
AT1G14960 |
AT1G14960
|
[Polyketide cyclase/dehydrase and lipid transport superfamily protein] |
-1.288 |
-0.211 |